9JN9
 
 | | Cryo-EM structure of human SLFN14 | | Descriptor: | Protein SLFN14, ZINC ION | | Authors: | Luo, M, Jia, X.D, Wang, Z.W, Yang, J.Y, Zhang, Q.F, Gao, S. | | Deposit date: | 2024-09-23 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and functional characterization of human SLFN14.
Nucleic Acids Res., 53, 2025
|
|
4X0U
 
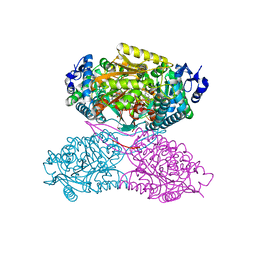 | | Structure ALDH7A1 inactivated by 4-diethylaminobenzaldehyde | | Descriptor: | 4-(diethylamino)benzaldehyde, Alpha-aminoadipic semialdehyde dehydrogenase, MAGNESIUM ION | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2014-11-23 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Diethylaminobenzaldehyde Is a Covalent, Irreversible Inactivator of ALDH7A1.
Acs Chem.Biol., 10, 2015
|
|
4X0T
 
 | |
6DWR
 
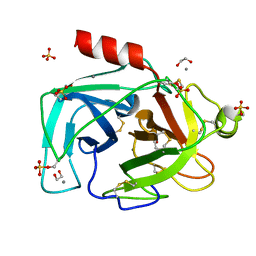 | | Trypsin serine protease modified with the protease inhibitor cyanobenzylsulfonylfluoride | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Luo, M, Eaton, C.N, Phillips-Piro, C.M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.319 Å) | | Cite: | Paired Spectroscopic and Crystallographic Studies of Protease Active Sites
Chemistryselect, 4, 2019
|
|
6DWQ
 
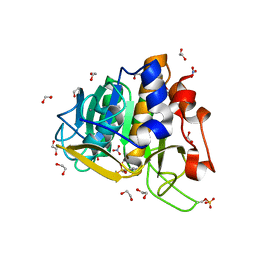 | | Subtilisin serine protease modified with the protease inhibitor cyanobenzylsulfonylfluoride | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, KerA, ... | | Authors: | Luo, M, Eaton, C.N, Phillips-Piro, C.M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Paired Spectroscopic and Crystallographic Studies of Protease Active Sites
Chemistryselect, 4, 2019
|
|
1TMF
 
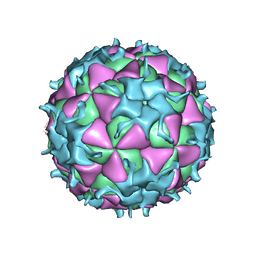 | | THREE-DIMENSIONAL STRUCTURE OF THEILER MURINE ENCEPHALOMYELITIS VIRUS (BEAN STRAIN) | | Descriptor: | THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP1), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP2), THEILER'S MURINE ENCEPHALOMYELITIS VIRUS (SUBUNIT VP3), ... | | Authors: | Luo, M, Toth, K.S. | | Deposit date: | 1992-02-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of Theiler murine encephalomyelitis virus (BeAn strain).
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
3L4N
 
 | | Crystal structure of yeast monothiol glutaredoxin Grx6 | | Descriptor: | GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Luo, M, Jiang, Y.-L, Ma, X.-X, He, Y.-X, Tang, Y.-J, Yu, J, Zhang, R.-G, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-04-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx6
J.Mol.Biol., 398, 2010
|
|
7L0X
 
 | |
7L0Y
 
 | |
7L0U
 
 | |
7L0V
 
 | |
7L0W
 
 | |
4ZUK
 
 | | Structure ALDH7A1 complexed with NAD+ | | Descriptor: | Alpha-aminoadipic semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TETRAETHYLENE GLYCOL | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
4ZUL
 
 | | Structure ALDH7A1 complexed with alpha-aminoadipate | | Descriptor: | 2-AMINOHEXANEDIOIC ACID, Alpha-aminoadipic semialdehyde dehydrogenase, TETRAETHYLENE GLYCOL, ... | | Authors: | Luo, M, Tanner, J.J. | | Deposit date: | 2015-05-16 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis of Substrate Recognition by Aldehyde Dehydrogenase 7A1.
Biochemistry, 54, 2015
|
|
3C8O
 
 | | The Crystal Structure of RraA from PAO1 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Regulator of ribonuclease activity A, ... | | Authors: | Luo, M, Niu, S, Yin, Y, Huang, A, Wang, D. | | Deposit date: | 2008-02-12 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of RraA from PAO1
To be published
|
|
4K79
 
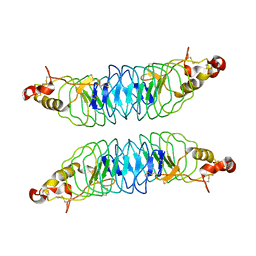 | | Recognition of the Thomsen-Friedenreich Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
4K5U
 
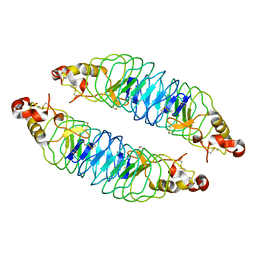 | | Recognition of the BG-H Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
6WTD
 
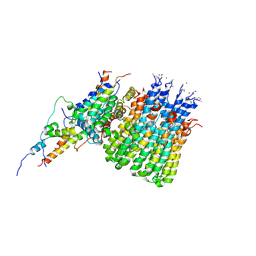 | | Monomer yeast ATP synthase Fo reconstituted in nanodisc with inhibitor of Bedaquiline bound | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Mueller, D.M, Srivastava, A.P, Symersky, J, Luo, M, Liao, M.F. | | Deposit date: | 2020-05-02 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Bedaquiline inhibits the yeast and human mitochondrial ATP synthases.
Commun Biol, 3, 2020
|
|
4ZCA
 
 | |
4ZHI
 
 | |
3PTX
 
 | |
3PTO
 
 | |
3PU4
 
 | |
3PU1
 
 | |
3PU0
 
 | |
