3CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN A347R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1AS4
 
 | | CLEAVED ANTICHYMOTRYPSIN A349R | | Descriptor: | ACETATE ION, ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
4CAA
 
 | | CLEAVED ANTICHYMOTRYPSIN T345R | | Descriptor: | ANTICHYMOTRYPSIN | | Authors: | Lukacs, C.M, Christianson, D.W. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering an anion-binding cavity in antichymotrypsin modulates the "spring-loaded" serpin-protease interaction.
Biochemistry, 37, 1998
|
|
1Z8D
 
 | | Crystal Structure of Human Muscle Glycogen Phosphorylase a with AMP and Glucose | | Descriptor: | ADENINE, ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, ... | | Authors: | Lukacs, C.M, Oikonomakos, N.G, Crowther, R.L, Hong, L.N, Kammlott, R.U, Levin, W, Li, S, Liu, C.M, Lucas-McGady, D, Pietranico, S, Reik, L. | | Deposit date: | 2005-03-30 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: An intermediate conformation with T-state and R-state features.
Proteins, 63, 2006
|
|
4WJB
 
 | | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Putative amidohydrolase/peptidase, SULFATE ION, ... | | Authors: | Lukacs, C.M, Dranow, D.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray crystal structure of a putative amidohydrolase/peptidase from Burkholderia cenocepacia
To Be Published
|
|
1D2I
 
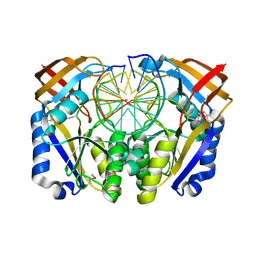 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | Descriptor: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
1DFM
 
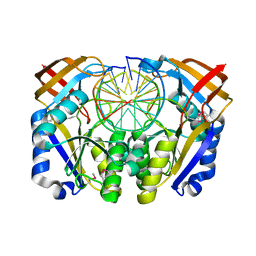 | | Crystal structure of restriction endonuclease BGLII complexed with DNA 16-mer | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
1ES8
 
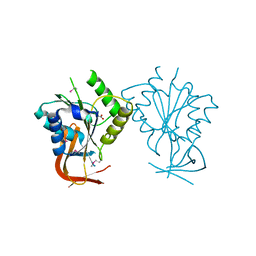 | | Crystal structure of free BglII | | Descriptor: | ACETIC ACID, RESTRICTION ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Aggarwal, A.K. | | Deposit date: | 2000-04-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease.
Nat.Struct.Biol., 8, 2001
|
|
4PN3
 
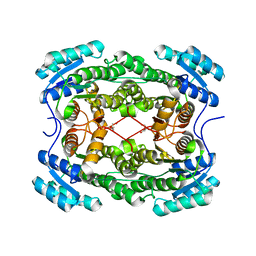 | | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyacyl-CoA dehydrogenase | | Authors: | Lukacs, C.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of 3-hydroxyacyl-CoA-dehydrogenase from Brucella melitensis
To Be Published
|
|
4E73
 
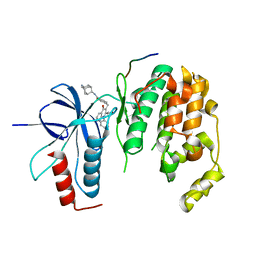 | | Crystal structure of JNK1beta-JIP in complex with an azaquinolone inhbitor | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, methyl 3-(4-{[(1R,2S,3S,5S,7s)-5-aminotricyclo[3.3.1.1~3,7~]dec-2-yl]carbamoyl}benzyl)-4-oxo-1-phenyl-1,4-dihydro-1,8-naphthyridine-2-carboxylate | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of an Adamantyl Azaquinolone JNK Selective Inhibitor.
ACS Med Chem Lett, 3, 2012
|
|
5W8J
 
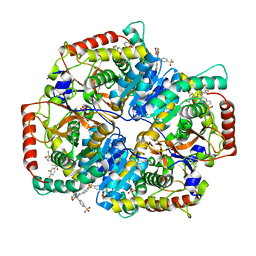 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | Descriptor: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lukacs, C.M, Moulin, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8K
 
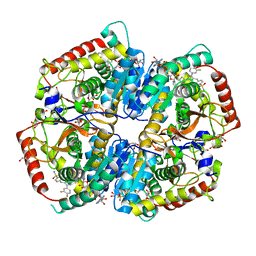 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8I
 
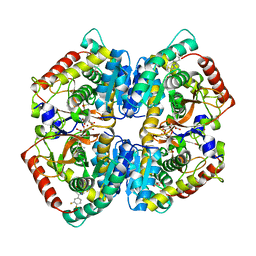 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8H
 
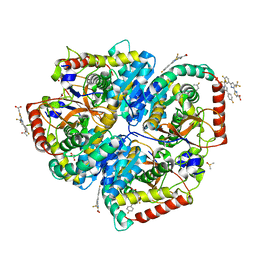 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 11 | | Descriptor: | 2-[3-(4-fluorophenyl)-5-(trifluoromethyl)-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
4KJU
 
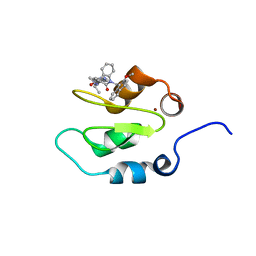 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
4KJV
 
 | | Crystal structure of XIAP-Bir2 with a bound spirocyclic benzoxazepinone inhibitor. | | Descriptor: | 6-methoxy-5-({(3S)-3-[(N-methyl-L-alanyl)amino]-4-oxo-2',3,3',4,5',6'-hexahydro-5H-spiro[1,5-benzoxazepine-2,4'-pyran]-5-yl}methyl)naphthalene-2-carboxylic acid, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
1P2A
 
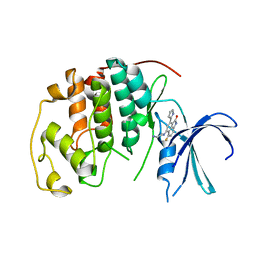 | | The structure of cyclin dependent kinase 2 (CKD2) with a trisubstituted naphthostyril inhibitor | | Descriptor: | 5-[(2-AMINOETHYL)AMINO]-6-FLUORO-3-(1H-PYRROL-2-YL)BENZO[CD]INDOL-2(1H)-ONE, Cell division protein kinase 2 | | Authors: | Liu, J.-J, Dermatakis, A, Lukacs, C.M, Konzelmann, F, Chen, Y, Kammlott, U, Depinto, W, Yang, H, Yin, X, Chen, Y, Schutt, A, Simcox, M.E, Luk, K.-C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3,5,6-Trisubstituted Naphthostyrils as CDK2 Inhibitors
BIOORG.MED.CHEM., 13, 2003
|
|
4NOZ
 
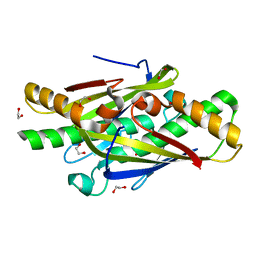 | | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Organic hydroperoxide resistance protein | | Authors: | Dranow, D.M, Lukacs, C.M, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Organic Hydroperoxide Resistance Protein from Burkholderia cenocepacia
TO BE PUBLISHED
|
|
3U15
 
 | | Structure of hDMX with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, Protein Mdm4, SULFATE ION | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VBG
 
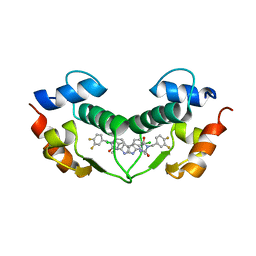 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LIH
 
 | |
4LFY
 
 | |
3ORN
 
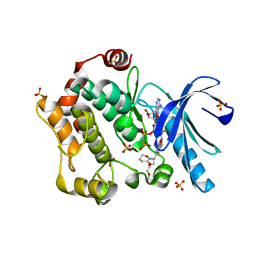 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4987655 and MgAMP-PNP | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(3-oxo-1,2-oxazinan-2-yl)methyl]benzamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OS3
 
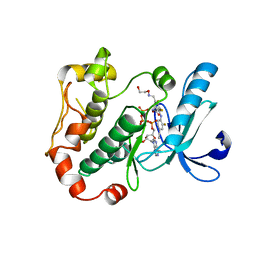 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4858061 and MgATP | | Descriptor: | 2-[(4-ethynyl-2-fluorophenyl)amino]-3,4-difluoro-N-(2-hydroxyethoxy)-5-{[(2-hydroxyethoxy)imino]methyl}benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3LE6
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a pyrazolobenzodiazepine inhibitor | | Descriptor: | 5-(2-chlorophenyl)-3-methyl-7-nitropyrazolo[3,4-b][1,4]benzodiazepine, Cell division protein kinase 2 | | Authors: | Lukacs, C.M, Swain, A, Crowther, R.L, Kammlott, R.U, Liu, J.J. | | Deposit date: | 2010-01-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolobenzodiazepines: part I. Synthesis and SAR of a potent class of kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
