1GLU
 
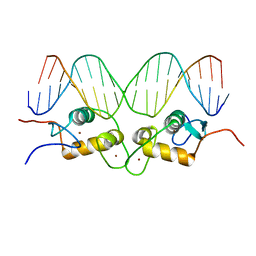 | | CRYSTALLOGRAPHIC ANALYSIS OF THE INTERACTION OF THE GLUCOCORTICOID RECEPTOR WITH DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*GP*AP*TP*GP*TP*TP*C P*TP*G)-3'), PROTEIN (GLUCOCORTICOID RECEPTOR), ZINC ION | | Authors: | Luisi, B.F, Xu, W.X, Otwinowski, Z, Freedman, L.P, Yamamoto, K.R, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA.
Nature, 352, 1991
|
|
1R4R
 
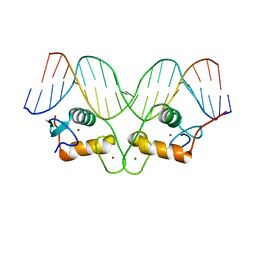 | | Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA | | Descriptor: | 5'-D(*CP*TP*GP*AP*GP*AP*AP*CP*AP*TP*CP*AP*TP*GP*TP*TP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*AP*AP*CP*AP*TP*GP*AP*TP*GP*TP*TP*CP*TP*CP*A)-3', Glucocorticoid receptor, ... | | Authors: | Luisi, B.F, Xu, W.X, Otwinowski, Z, Freedman, L.P, Yamamoto, K.R, Sigler, P.B. | | Deposit date: | 2003-10-07 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic Analysis of the Interaction of the Glucocorticoid Receptor with DNA
Nature, 352, 1991
|
|
1R4O
 
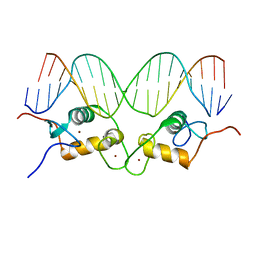 | | Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*GP*AP*TP*GP*TP*TP*CP*TP*G)-3', Glucocorticoid receptor, ZINC ION | | Authors: | Luisi, B.F, Xu, W.X, Otwinowski, Z, Freedman, L.P, Yamamoto, K.R, Sigler, P.B. | | Deposit date: | 2003-10-07 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Analysis of the Interaction of The Glucocorticoid Receptor with DNA
Nature, 352, 1991
|
|
7PCR
 
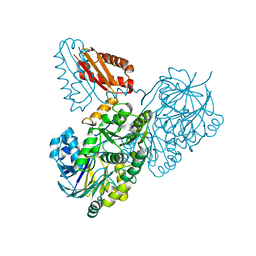 | | Helicobacter pylori RNase J | | Descriptor: | Ribonuclease J | | Authors: | Luisi, B.F, Pei, X.Y. | | Deposit date: | 2021-08-03 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Acetylation regulates the oligomerization state and activity of RNase J, the Helicobacter pylori major ribonuclease.
Nat Commun, 14, 2023
|
|
5NQB
 
 | |
5NQQ
 
 | |
8B0J
 
 | | CryoEM structure of bacterial RNaseE.RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small RNA, RNase adapter protein RapZ, Ribonuclease E | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8B0I
 
 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
4V2S
 
 | | Crystal structure of Hfq in complex with the sRNA RydC | | Descriptor: | RNA-BINDING PROTEIN HFQ, RYDC | | Authors: | Dimastrogiovanni, D, Frohlich, K.S, Bruce, H.A, Bandyra, K.J, Hohensee, S, Vogel, J, Luisi, B.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Recognition of the small regulatory RNA RydC by the bacterial Hfq protein.
Elife, 3, 2014
|
|
1TRO
 
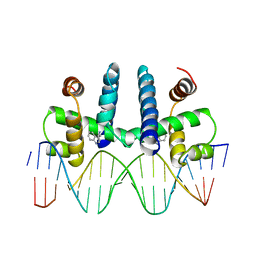 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
9QH3
 
 | |
9QH0
 
 | |
1W85
 
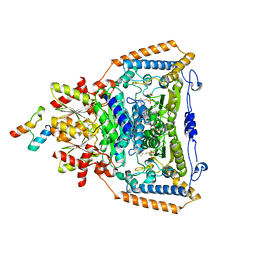 | | The crystal structure of pyruvate dehydrogenase E1 bound to the peripheral subunit binding domain of E2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A molecular switch and proton wire synchronize the active sites in thiamine enzymes.
Science, 306, 2004
|
|
1W88
 
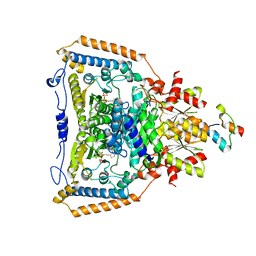 | | The crystal structure of pyruvate dehydrogenase E1(D180N,E183Q) bound to the peripheral subunit binding domain of E2 | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE COMPONENT OF PYRUVATE, MAGNESIUM ION, PYRUVATE DEHYDROGENASE E1 COMPONENT, ... | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Switch and Proton-Wire Synchronize the Active Sites in Thiamine-Dependent Enzymes
Science, 306, 2004
|
|
6G63
 
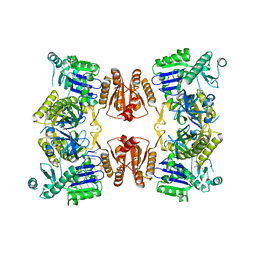 | | RNase E in complex with sRNA RrpA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), Ribonuclease E, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Bandyra, K.B, Luisi, B.F. | | Deposit date: | 2018-03-31 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate Recognition and Autoinhibition in the Central Ribonuclease RNase E.
Mol. Cell, 72, 2018
|
|
6GWK
 
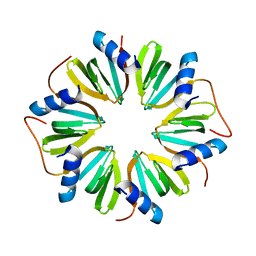 | | The crystal structure of Hfq from Caulobacter crescentus | | Descriptor: | RNA-binding protein Hfq | | Authors: | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HCK
 
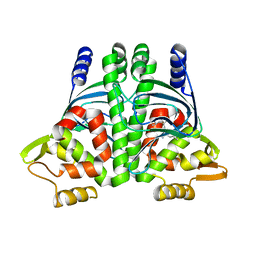 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with dipeptide Leu-Leu | | Descriptor: | LEUCINE, Listeriolysin regulatory protein, SODIUM ION | | Authors: | Grundstrom, C, Oelker, M, Krypotou, E, Scortti, M, Luisi, B.F, Vazquez-Boland, J, Sauer-Eriksson, A.E. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of Bacterial Virulence through the Peptide Signature of the Habitat.
Cell Rep, 26, 2019
|
|
2V07
 
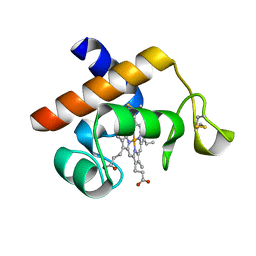 | | Structure of the Arabidopsis thaliana cytochrome c6A V52Q variant | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
2VMK
 
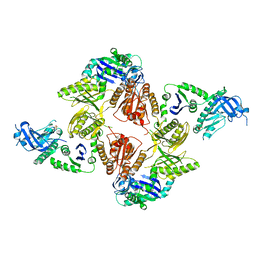 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2V08
 
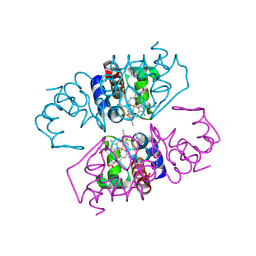 | | Structure of wild-type Phormidium laminosum cytochrome c6 | | Descriptor: | CHLORIDE ION, CYTOCHROME C6, HEME C, ... | | Authors: | Worrall, J.A.R, Schlarb-Ridley, B.G, Reda, T, Marcaida, M.J, Moorlen, R.J, Wastl, J, Hirst, J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2007-05-10 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modulation of heme redox potential in the cytochrome c6 family.
J. Am. Chem. Soc., 129, 2007
|
|
8CGL
 
 | |
3DV0
 
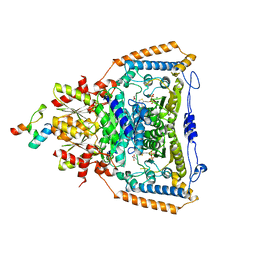 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DVA
 
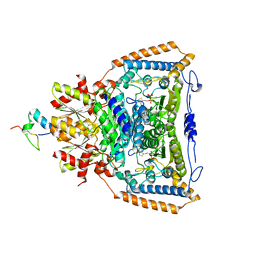 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DUF
 
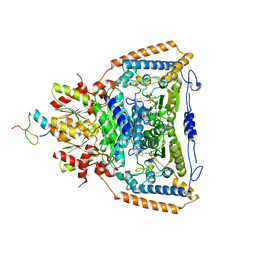 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
7AUA
 
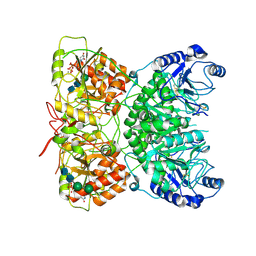 | | Cryo-EM structure of human exostosin-like 3 (EXTL3) in complex with UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Wilson, L.F.L, Dendooven, T, Hardwick, S.W, Chirgadze, D.Y, Luisi, B.F, Logan, D.T, Mani, K, Dupree, P. | | Deposit date: | 2020-11-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The structure of EXTL3 helps to explain the different roles of bi-domain exostosins in heparan sulfate synthesis.
Nat Commun, 13, 2022
|
|
