1JJS
 
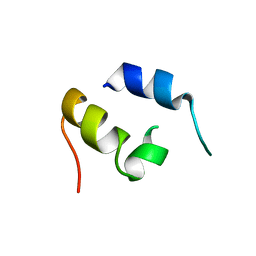 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
2ARG
 
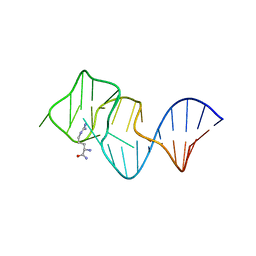 | | FORMATION OF AN AMINO ACID BINDING POCKET THROUGH ADAPTIVE ZIPPERING-UP OF A LARGE DNA HAIRPIN LOOP, NMR, 9 STRUCTURES | | Descriptor: | ARGININEAMIDE, DNA APTAMER [5'-D (*TP*GP*AP*CP*CP*AP*GP*GP*GP*CP*AP*AP*AP*CP*GP*GP*TP*AP* GP*GP*TP*GP*AP*GP*TP*GP*GP*TP*CP*A)-3'] | | Authors: | Lin, C.H, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-08-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Formation of an amino-acid-binding pocket through adaptive zippering-up of a large DNA hairpin loop.
Chem.Biol., 5, 1998
|
|
1OLD
 
 | |
107D
 
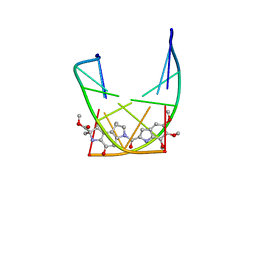 | | SOLUTION STRUCTURE OF THE COVALENT DUOCARMYCIN A-DNA DUPLEX COMPLEX | | Descriptor: | 4-HYDROXY-2,8-DIMETHYL-1-OXO-6-(4,5,6-TRIMETHOXY-1H-INDOLE-2-CARBONYL)-1,2,3,6,7,8-HEXAHYDRO-3,6-DIAZA-AS-INDACENE-2-CARBOXYLIC ACID METHYL ESTER, DNA (5'-D(*CP*CP*TP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*AP*GP*G)-3') | | Authors: | Lin, C.H, Patel, D.J. | | Deposit date: | 1995-01-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the covalent duocarmycin A-DNA duplex complex.
J.Mol.Biol., 248, 1995
|
|
1AW4
 
 | |
1HWV
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1S)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-10 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
1HX4
 
 | | MOLECULAR TOPOLOGY OF POLYCYCLIC AROMATIC CARCINOGENS DETERMINES DNA ADDUCT CONFORMATION: A LINK TO TUMORIGENIC ACTIVITY | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, 5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Patel, D.J, Lin, C.H, Geacintov, N.E, Broyde, S, Huang, X, Kolbanovskii, A, Hingerty, B.E, Amin, S. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity.
J.Mol.Biol., 306, 2001
|
|
4Y24
 
 | | Complex of human Galectin-1 and TD-139 | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Dual thio-digalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors.
Sci Rep, 6, 2016
|
|
4XBN
 
 | |
4XBQ
 
 | |
4Y1V
 
 | | Complex of human Galectin-1 and Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1U
 
 | | Complex of human Galectin-1 and Galbeta1-4GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y26
 
 | | Complex of human Galectin-7 and Galbeta1-3(6OSO3)GlcNAc | | Descriptor: | Galectin-7, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1Z
 
 | | Complex of human Galectin-1 and Galbeta1-4(6CO2)GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranosiduronic acid | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y22
 
 | | Complex of human Galectin-1 and (3OSO3)Galbeta1-3GlcNAc | | Descriptor: | 3-O-sulfo-beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Galectin-1 | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y1Y
 
 | | Complex of human Galectin-1 and (6OSO3)Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4Y20
 
 | | Complex of human Galectin-1 and NeuAcalpha2-3Galbeta1-4Glc | | Descriptor: | Galectin-1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
4XBL
 
 | |
4Y1X
 
 | | Complex of human Galectin-1 and Galbeta1-4(6OSO3)GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-4)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
5Z19
 
 | |
5Z1A
 
 | |
5Z1B
 
 | |
5Z18
 
 | |
9IIS
 
 | | GDP-fucose pyrophosphorylase part of FKP with a SUMO tag | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Ubiquitin-like protein SMT3,L-fucokinase/L-fucose-1-P guanylyltransferase | | Authors: | Ko, T.P, Lin, S.W, Hsu, M.F, Lin, C.H. | | Deposit date: | 2024-06-21 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural insight into the catalytic mechanism of the bifunctional enzyme l-fucokinase/GDP-fucose pyrophosphorylase.
J.Biol.Chem., 301, 2025
|
|
9IIP
 
 | | Fucokinase part of FKP with beta-L-fucose 1-phosphate | | Descriptor: | 6-deoxy-1-O-phosphono-beta-L-galactopyranose, L-fucokinase/L-fucose-1-P guanylyltransferase | | Authors: | Ko, T.P, Lin, S.W, Hsu, M.F, Lin, C.H. | | Deposit date: | 2024-06-21 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into the catalytic mechanism of the bifunctional enzyme l-fucokinase/GDP-fucose pyrophosphorylase.
J.Biol.Chem., 301, 2025
|
|
