1FAS
 
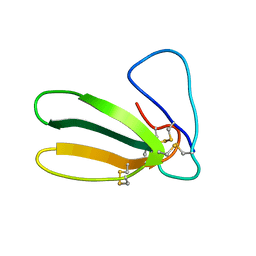 | | 1.9 ANGSTROM RESOLUTION STRUCTURE OF FASCICULIN 1, AN ANTI-ACETYLCHOLINESTERASE TOXIN FROM GREEN MAMBA SNAKE VENOM | | Descriptor: | FASCICULIN 1 | | Authors: | Le Du, M.H, Marchot, P, Bougis, P.E, Fontecilla-Camps, J.C. | | Deposit date: | 1992-08-07 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.9-A resolution structure of fasciculin 1, an anti-acetylcholinesterase toxin from green mamba snake venom.
J.Biol.Chem., 267, 1992
|
|
1QM7
 
 | | X-ray structure of a three-fingered chimeric protein, stability of a structural scaffold | | Descriptor: | R-CHII | | Authors: | Le Du, M.H, Ricciardi, A, Khayati, M, Menez, R, Boulain, J.C, Menez, A, Ducancel, F. | | Deposit date: | 1999-09-21 | | Release date: | 2000-03-15 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of a Structural Scaffold Upon Activity Transfer : X-Ray Structure of a Three Fingers Chimeric Protein.
J.Mol.Biol., 296, 2000
|
|
1EW2
 
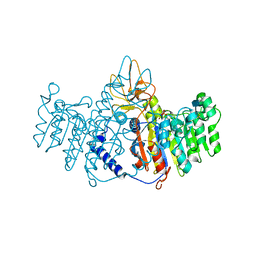 | | CRYSTAL STRUCTURE OF A HUMAN PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, PHOSPHATASE, ... | | Authors: | Le Du, M.H, Stigbrand, T, Taussig, M.J, Menez, A, Stura, E.A. | | Deposit date: | 2000-04-21 | | Release date: | 2001-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of alkaline phosphatase from human placenta at 1.8 A resolution. Implication for a substrate specificity.
J.Biol.Chem., 276, 2001
|
|
1KH9
 
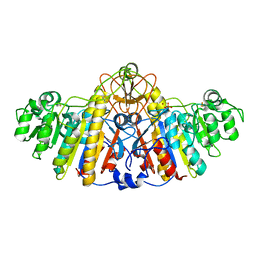 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHK
 
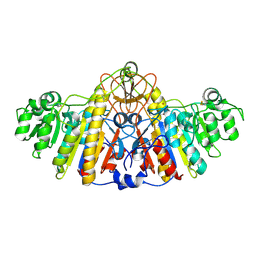 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) | | Descriptor: | Alkaline Phosphatase, MAGNESIUM ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH7
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153GD330N) | | Descriptor: | MAGNESIUM ION, SULFATE ION, ZINC ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHL
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) COMPLEX WITH PHOSPHATE | | Descriptor: | Alkaline Phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHN
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH4
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH5
 
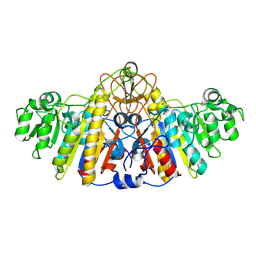 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
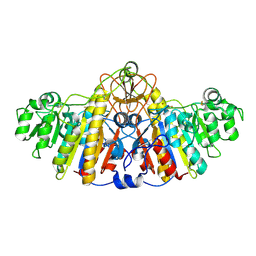 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1FSC
 
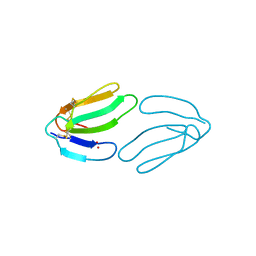 | |
7BDX
 
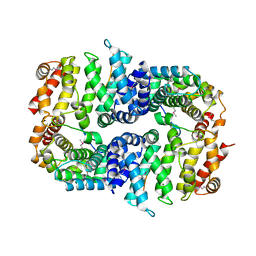 | | Armadillo domain of HSF2BP in complex with BRCA2 peptide | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein, MAGNESIUM ION | | Authors: | Le Du, M.H, Zinn-Justin, S, Ghouil, R, Miron, S, Legrand, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-07 | | Last modified: | 2022-07-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BRCA2 binding through a cryptic repeated motif to HSF2BP oligomers does not impact meiotic recombination.
Nat Commun, 12, 2021
|
|
1RCN
 
 | | CRYSTAL STRUCTURE OF THE RIBONUCLEASE A D(APTPAPAPG) COMPLEX : DIRECT EVIDENCE FOR EXTENDED SUBSTRATE RECOGNITION | | Descriptor: | DNA (5'-D(*AP*TP*AP*A)-3'), PROTEIN (RIBONUCLEASE A (E.C.3.1.27.5)) | | Authors: | Fontecilla-Camps, J.C, De Llorens, R, Le Du, M.H, Cuchillo, C.M. | | Deposit date: | 1994-05-27 | | Release date: | 1994-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition.
J.Biol.Chem., 269, 1994
|
|
1YNT
 
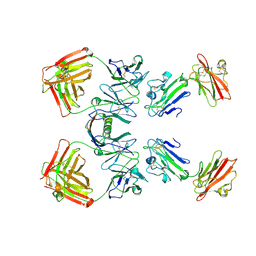 | | Structure of the monomeric form of T. gondii SAG1 surface antigen bound to a human Fab | | Descriptor: | 4F11E12 Fab variable heavy chain region, 4F11E12 Fab variable light chain region, CADMIUM ION, ... | | Authors: | Graille, M, Stura, E.A, Bossus, M, Muller, B.H, Letourneur, O, Battail-Poirot, N, Sibai, G, Rolland, D, Le Du, M.H, Ducancel, F. | | Deposit date: | 2005-01-25 | | Release date: | 2005-12-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the complex between the monomeric form of Toxoplasma gondii surface antigen 1 (SAG1) and a monoclonal antibody that mimics the human immune response
J.Mol.Biol., 354, 2005
|
|
1ZEB
 
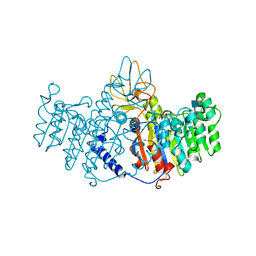 | | X-ray structure of alkaline phosphatase from human placenta in complex with 5'-AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1YWH
 
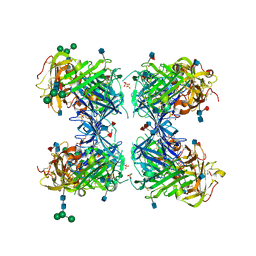 | | crystal structure of urokinase plasminogen activator receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Llinas, P, Le Du, M.H, Gardsvoll, H, Dano, K, Ploug, M, Gilquin, B, Stura, E.A, Menez, A. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide
EMBO J., 24, 2005
|
|
1ZED
 
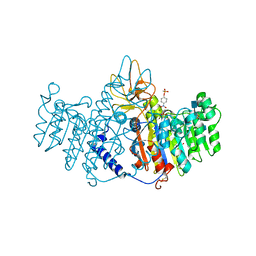 | | Alkaline phosphatase from human placenta in complex with p-nitrophenyl-phosphonate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1ZEF
 
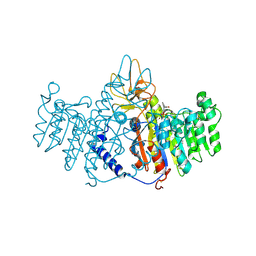 | | structure of alkaline phosphatase from human placenta in complex with its uncompetitive inhibitor L-Phe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, CALCIUM ION, ... | | Authors: | Llinas, P, Stura, E.A, Menez, A, Kiss, Z, Stigbrand, T, Millan, J.L, Le Du, M.H. | | Deposit date: | 2005-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Human Placental Alkaline Phosphatase in Complex with Functional Ligands.
J.Mol.Biol., 350, 2005
|
|
1FF4
 
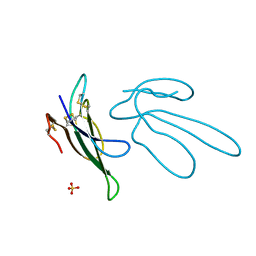 | |
1GZF
 
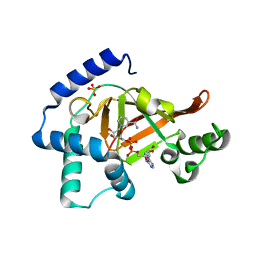 | | Structure of the Clostridium botulinum C3 exoenzyme (wild-type) in complex with NAD | | Descriptor: | 3-(AMINOCARBONYL)-1-[(3R,4S,5R)-3,4-DIHYDROXY-5-METHYLTETRAHYDRO-2-FURANYL]PYRIDINIUM, ADENOSINE-5'-DIPHOSPHATE, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Menetrey, J, Flatau, G, Stura, E.A, Charbonnier, J.B, Gas, F, Teulon, J.M, Le Du, M.H, Boquet, P, Menez, A. | | Deposit date: | 2002-05-21 | | Release date: | 2002-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nad Binding Induces Conformational Changes in Rho Adp-Ribosylating Clostridium Botulinum C3 Exoenzyme
J.Biol.Chem., 277, 2002
|
|
2H7Z
 
 | | Crystal structure of irditoxin | | Descriptor: | Irditoxin subunit A, Irditoxin subunit B | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-29 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity.
Faseb J., 23, 2009
|
|
2GLQ
 
 | | X-ray structure of human alkaline phosphatase in complex with strontium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, placental type, ... | | Authors: | Llinas, P, Masella, M, Stigbrand, T, Menez, A, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of human alkaline phosphatase in complex with strontium: Implication for its secondary effect in bones.
Protein Sci., 15, 2006
|
|
2HEO
 
 | | General Structure-Based Approach to the Design of Protein Ligands: Application to the Design of Kv1.2 Potassium Channel Blockers. | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Z-DNA binding protein 1 | | Authors: | Magis, C, Gasparini, S, Charbonnier, J.B, Stura, E, Le Du, M.H, Menez, A, Cuniasse, P. | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based secondary structure-independent approach to design protein ligands: Application to the design of Kv1.2 potassium channel blockers.
J.Am.Chem.Soc., 128, 2006
|
|
3G5H
 
 | | Crystallographic analysis of cytochrome P450 cyp121 | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Belin, P, Le Du, M.H, Gondry, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
