6MCW
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, complex with the detergent Anapoe-X-114 | | Descriptor: | 23-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]-3,6,9,12,15,18,21-heptaoxatricosan-1-ol, Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6MI0
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, ligand-free state | | Descriptor: | Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-18 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
2BN4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2005-03-18 | | Release date: | 2006-01-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
2BF4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
7JJ1
 
 | |
1SE6
 
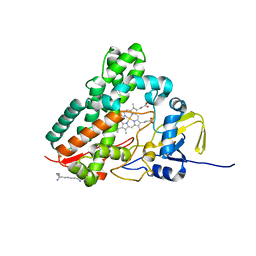 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SPERMINE (FULLY PROTONATED FORM), ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-02-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
5M5H
 
 | | RIBOSOME-BOUND YIDC INSERTASE | | Descriptor: | Membrane protein insertase YidC | | Authors: | Kedrov, A, Wickles, S, Crevenna, A.H, van der Sluis, E, Buschauer, R, Berninghausen, O, Lamb, D.C, Beckmann, R. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.
Cell Rep, 17, 2016
|
|
1ODO
 
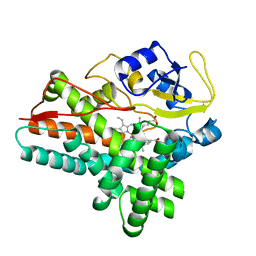 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
1S1F
 
 | | Crystal Structure of Streptomyces Coelicolor A3(2) CYP158A2 from antibiotic biosynthetic pathways | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, GLYCEROL, MALONIC ACID, ... | | Authors: | Zhao, B, Lamb, D.C, Lei, L, Sundaramoorthy, M, Podust, L.M, Waterman, M.R. | | Deposit date: | 2004-01-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2
J.Biol.Chem., 280, 2005
|
|
1GWI
 
 | | The 1.92 A structure of Streptomyces coelicolor A3(2) CYP154C1: A new monooxygenase that functionalizes macrolide ring systems | | Descriptor: | CYTOCHROME P450 154C1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Podust, L.M, Kim, Y, Arase, M, Neely, B.A, Beck, B.J, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2002-03-15 | | Release date: | 2003-01-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.92 A Structure of Streptomyces Coelicolor A3(2) Cyp154C1: A New Monooxygenase that Functionalizes Macrolide Ring Systems
J.Biol.Chem., 278, 2003
|
|
4B8N
 
 | | Cytochrome b5 of Ostreococcus tauri virus 2 | | Descriptor: | CYTOCHROME B5-HOST ORIGIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Isupov, M, Reid, E.L, Weynberg, K.D, Love, J, Wilson, W.H, Kelly, S.L, Lamb, D.C, Allen, M.J, Littlechild, J.A. | | Deposit date: | 2012-08-28 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Characterisation of a Viral Cytochrome B5.
FEBS Lett., 587, 2013
|
|
2NZ5
 
 | | Structure and Function Studies of Cytochrome P450 158A1 from Streptomyces coelicolor A3(2) | | Descriptor: | Cytochrome P450 CYP158A1, PROTOPORPHYRIN IX CONTAINING FE, naphthalene-1,2,4,5,7-pentol | | Authors: | Zhao, B, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2006-11-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2.
Biochemistry, 46, 2007
|
|
6N6Q
 
 | |
4UYM
 
 | |
4UYL
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51B) from a pathogenic filamentous fungus Aspergillus fumigatus in complex with VNI | | Descriptor: | 14-ALPHA STEROL DEMETHYLASE, N-[(1R)-1-(2,4-dichlorophenyl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T.Y, Wawrzak, Z, Lepesheva, G.I. | | Deposit date: | 2014-09-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-Functional Characterization of Cytochrome P450 Sterol Alpha-Demethylase (Cyp51B) from Aspergillus Fumigatus and Molecular Basis for the Development of Antifungal Drugs
J.Biol.Chem., 290, 2015
|
|
1T93
 
 | | Evidence for Multiple Substrate Recognition and Molecular Mechanism of C-C reaction by Cytochrome P450 CYP158A2 from Streptomyces Coelicolor A3(2) | | Descriptor: | FLAVIOLIN, PROTOPORPHYRIN IX CONTAINING FE, putative cytochrome P450 | | Authors: | Zhao, B, Sundaramoorthy, M, Waterman, M.R. | | Deposit date: | 2004-05-14 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Binding of Two Flaviolin Substrate Molecules, Oxidative Coupling, and Crystal Structure of Streptomyces coelicolor A3(2) Cytochrome P450 158A2.
J.Biol.Chem., 280, 2005
|
|
7NDX
 
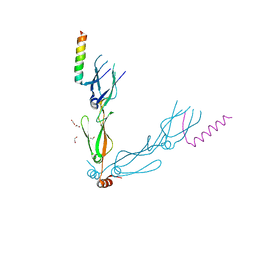 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | Descriptor: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | Authors: | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
7PKP
 
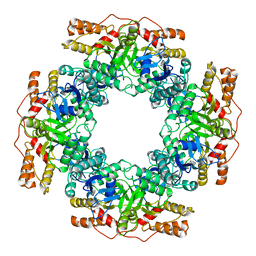 | | NSP2 RNP complex | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PKO
 
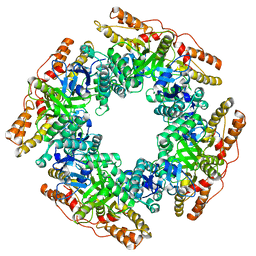 | | CryoEM structure of Rotavirus NSP2 | | Descriptor: | Non-structural protein 2 | | Authors: | Bravo, J.P.K, Borodavka, A. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of rotavirus RNA chaperone displacement and RNA annealing.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EU7
 
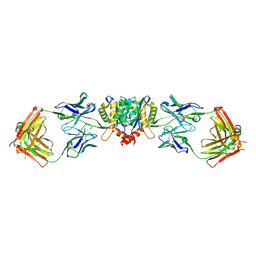 | | Crystal structure of HIV-1 integrase catalytic core in complex with Fab | | Descriptor: | FAB Heavy Chain, FAB light chain, Integrase | | Authors: | Galilee, M, Griner, S.L, Stroud, R.M, Alian, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Preserved HTH-Docking Cleft of HIV-1 Integrase Is Functionally Critical.
Structure, 24, 2016
|
|
8FZ8
 
 | | Structure of cytochrome P450sky2 | | Descriptor: | Cytochrome P450, OCTANOIC ACID (CAPRYLIC ACID), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murarka, V.C, Poulos, T.L. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Biosynthesis of a new skyllamycin in Streptomyces nodosus : a cytochrome P450 forms an epoxide in the cinnamoyl chain.
Org.Biomol.Chem., 22, 2024
|
|
3DBG
 
 | |
3EL3
 
 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
2DKK
 
 | |
7SHI
 
 | | Crystal Structure of Cytochrome P450 AmphL from Streptomyces nodosus and the Structural Basis for Substrate Selectivity in Macrolide Metabolizing P450s | | Descriptor: | (1R,3S,5R,6R,9R,11R,15S,16R,17R,18S,19E,22E,33S,35S,36R,37S)-33-[(3-amino-3,6-dideoxy-beta-D-mannopyranosyl)oxy]-1,3,5,6,9,11,17,37-octahydroxy-15,16,18-trimethyl-13-oxo-14,39-dioxabicyclo[33.3.1]nonatriaconta-19,22-diene-36-carboxylic acid, AmphL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Amaya, J.A, Poulos, T.L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.00002766 Å) | | Cite: | Structural analysis of P450 AmphL from Streptomyces nodosus provides insights into substrate selectivity of polyene macrolide antibiotic biosynthetic P450s.
J.Biol.Chem., 298, 2022
|
|
