1NTP
 
 | |
1TGN
 
 | |
3PGF
 
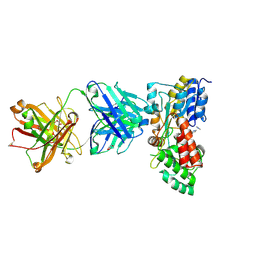 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
8RQF
 
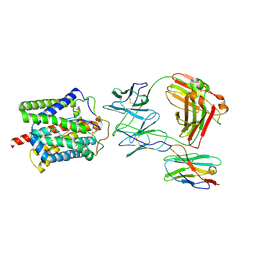 | | Cryo-EM structure of human NTCP-Bulevirtide complex | | Descriptor: | Fab-specific nanobody, Heavy chain of Fab3, Light chain of Fab3, ... | | Authors: | Liu, H, Zakrzewicz, D, Nosol, K, Irobalieva, R.N, Mukherjee, S, Bang-Soerensen, R, Goldmann, N, Kunz, S, Rossi, L, Kossiakoff, A.A, Urban, S, Glebe, D, Geyer, J, Locher, K.P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP.
Nat Commun, 15, 2024
|
|
5UEA
 
 | |
7RTH
 
 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHP
 
 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PIJ
 
 | | Structure of Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMTNb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHQ
 
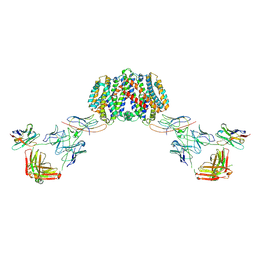 | | Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMT-Nb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9AVO
 
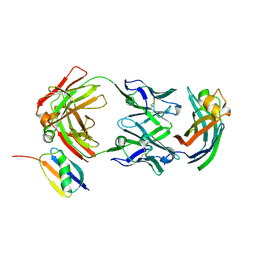 | |
9AWE
 
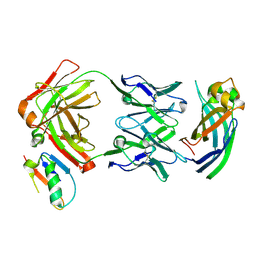 | |
5CWW
 
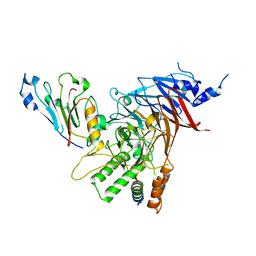 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWV
 
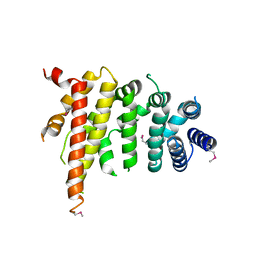 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWU
 
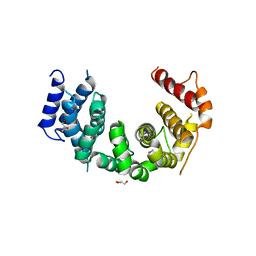 | | Crystal structure of Chaetomium thermophilum Nup188 TAIL domain | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5UCB
 
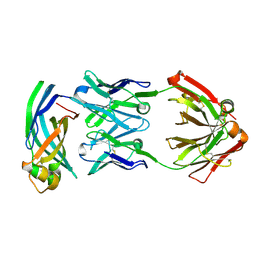 | |
3BOG
 
 | | Snow Flea Antifreeze Protein Quasi-racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein, UNKNOWN LIGAND | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
3BOI
 
 | | Snow Flea Antifreeze Protein Racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
7ZLH
 
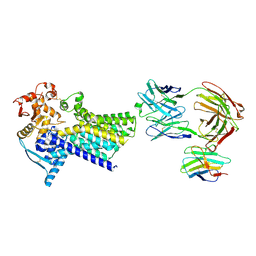 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLG
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZYI
 
 | | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | | Descriptor: | CHOLESTEROL, GLYCOCHENODEOXYCHOLIC ACID, Nanobody, ... | | Authors: | Liu, H, Irobalieva, R.N, Bang-Sorensen, R, Nosol, K, Mukherjee, S, Agrawal, P, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver.
Cell Res., 32, 2022
|
|
8PG0
 
 | | Human OATP1B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-17 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
8PHW
 
 | | Human OATP1B1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Fab18 (heavy chain, variable region), ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
6RLX
 
 | |
