5I08
 
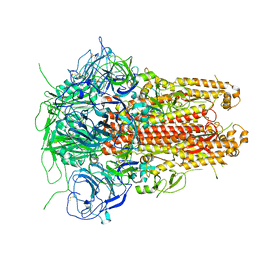 | | Prefusion structure of a human coronavirus spike protein | | Descriptor: | Spike glycoprotein,Foldon chimera | | Authors: | Kirchdoerfer, R.N, Cottrell, C.A, Wang, N, Pallesen, J, Yassine, H.M, Turner, H.L, Corbett, K.S, Graham, B.S, McLellan, J.S, Ward, A.B. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pre-fusion structure of a human coronavirus spike protein.
Nature, 531, 2016
|
|
3QG6
 
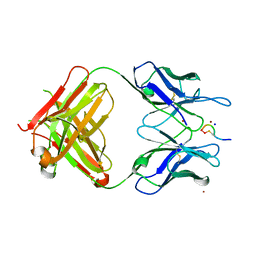 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Heavy Chain, AP4-24H11 Light Chain, Agr autoinducing peptide, ... | | Authors: | Kirchdoerfer, R.N, Janda, J.D, Kaufmann, G.F, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
6VV5
 
 | |
6NUR
 
 | |
6NUS
 
 | |
6NUT
 
 | | Ebola virus nucleoprotein - RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3') | | Authors: | Kirchdoerfer, R.N, Ward, A.B. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4ZTG
 
 | |
4ZTI
 
 | |
4ZTA
 
 | |
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | Descriptor: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
3TYJ
 
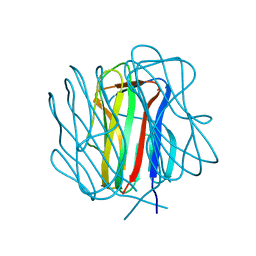 | | Bacillus collagen-like protein of anthracis P159S mutant | | Descriptor: | BclA protein | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
5T3T
 
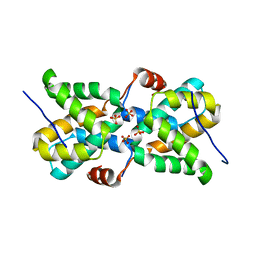 | | Ebola virus VP30 CTD bound to nucleoprotein | | Descriptor: | Fusion protein of Nucleoprotein and Minor nucleoprotein VP30, SULFATE ION | | Authors: | Kirchdoerfer, R.N, Moyer, C.L, Abelson, D.M, Saphire, E.O. | | Deposit date: | 2016-08-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis.
Plos Pathog., 12, 2016
|
|
6CS2
 
 | | SARS Spike Glycoprotein - human ACE2 complex, Stabilized variant, all ACE2-bound particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS1
 
 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, two S1 CTDs in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRZ
 
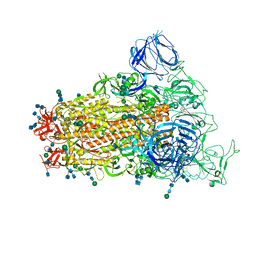 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CS0
 
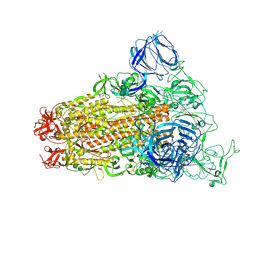 | | SARS Spike Glycoprotein, Trypsin-cleaved, Stabilized variant, one S1 CTD in an upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRX
 
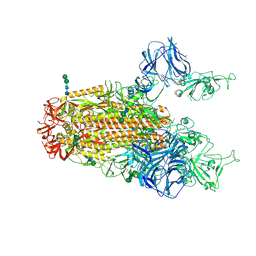 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRW
 
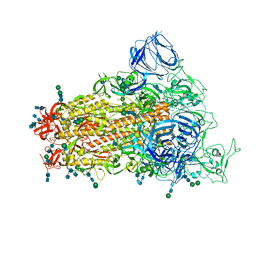 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CRV
 
 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
8G6R
 
 | | Porcine epidemic diarrhea virus core polymerase complex | | Descriptor: | RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3'), RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3'), ZINC ION, ... | | Authors: | Anderson, T.K, Kirchdoerfer, R.N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-03-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An alphacoronavirus polymerase structure reveals conserved co-factor functions.
Biorxiv, 2023
|
|
8URB
 
 | |
2YMN
 
 | | Organization of the Influenza Virus Replication Machinery | | Descriptor: | NUCLEOPROTEIN | | Authors: | Moeller, A, Kirchdoerfer, R.N, Potter, C.S, Carragher, B, Wilson, I.A. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Organization of the Influenza Virus Replication Machinery.
Science, 338, 2012
|
|
9CPO
 
 | |
7LHQ
 
 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
