2MRA
 
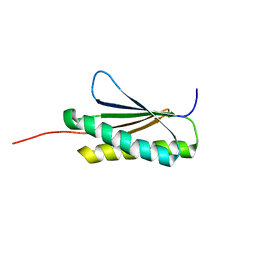 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | Descriptor: | De novo designed protein OR459 | | Authors: | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
3TC6
 
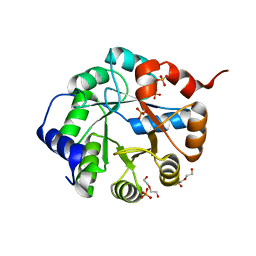 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TC7
 
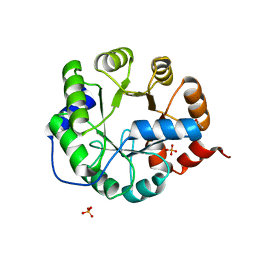 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
4LNY
 
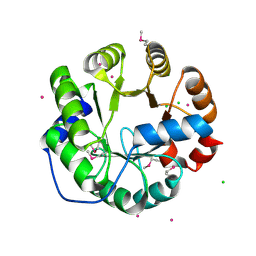 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR422 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Engineered Protein OR422 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Maglaqui, M, Kogan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Crystal Structure of Engineered Protein OR422.
To be Published
|
|
4LT9
 
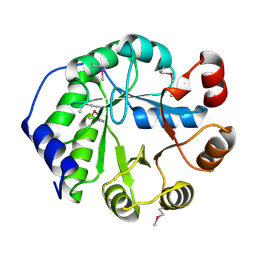 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
8AH9
 
 | |
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMX
 
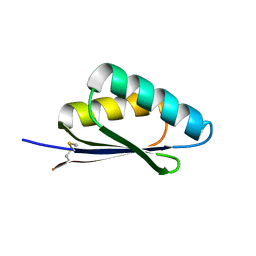 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
4OU1
 
 | | Crystal structure of a computationally designed retro-aldolase covalently bound to folding probe 1 [(6-methoxynaphthalen-2-yl)(oxiran-2-yl)methanol] | | Descriptor: | (1S,2S)-1-(6-methoxynaphthalen-2-yl)propane-1,2-diol, BENZOIC ACID, PHOSPHATE ION, ... | | Authors: | Bhabha, G, Zhang, X, Ekiert, D.C. | | Deposit date: | 2014-02-14 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3T1G
 
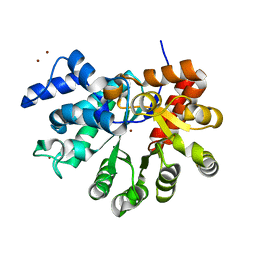 | |
8U5A
 
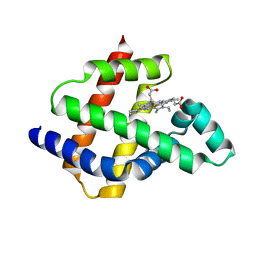 | | Improving protein expression, stability, and function with ProteinMPNN | | Descriptor: | Designed myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kalvet, I, Bera, A.K, Baker, D. | | Deposit date: | 2023-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving Protein Expression, Stability, and Function with ProteinMPNN.
J.Am.Chem.Soc., 146, 2024
|
|
8TCG
 
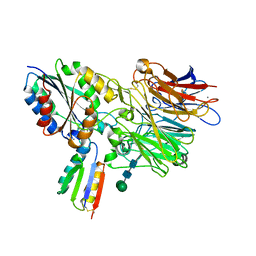 | | Integrin alpha-v beta-6 in complex with minibinder B6_BP_dslf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Integrin alpha-V heavy chain, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
8TCF
 
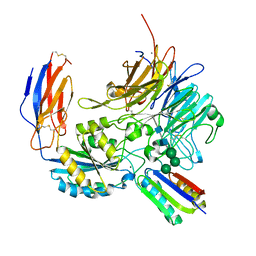 | | Integrin alpha-v beta-8 in complex with minibinder B8_BP_dsulf | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Campbell, M.G, Fernandez, A, Roy, A, Kraft, J, Baker, D. | | Deposit date: | 2023-06-30 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
4PEK
 
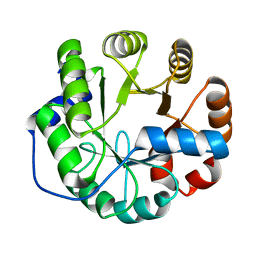 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEJ
 
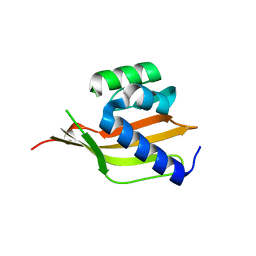 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | Descriptor: | Retro-aldolase | | Authors: | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
