2HDC
 
 | | STRUCTURE OF TRANSCRIPTION FACTOR GENESIS/DNA COMPLEX | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*AP*AP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*TP*TP*AP*AP*GP*C)-3'), PROTEIN (TRANSCRIPTION FACTOR) | | Authors: | Jin, C, Marsden, I, Chen, X, Liao, X. | | Deposit date: | 1999-05-05 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex.
J.Mol.Biol., 289, 1999
|
|
2HO9
 
 | |
2MOK
 
 | | holo_FldA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2014-04-27 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR study of YqcA from Escherichia coli
To be Published
|
|
8XEM
 
 | |
8XEO
 
 | |
8XDJ
 
 | |
8XEH
 
 | |
8YUF
 
 | |
8YOT
 
 | |
2IPA
 
 | | solution structure of Trx-ArsC complex | | Descriptor: | Protein arsC, Thioredoxin | | Authors: | Jin, C, Hu, Y, Li, Y, Zhang, X. | | Deposit date: | 2006-10-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conformational fluctuations coupled to the thiol-disulfide transfer between thioredoxin and arsenate reductase in Bacillus subtilis.
J.Biol.Chem., 282, 2007
|
|
5B7J
 
 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
2OQ3
 
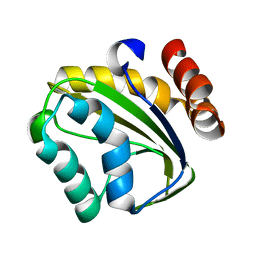 | |
1Z2D
 
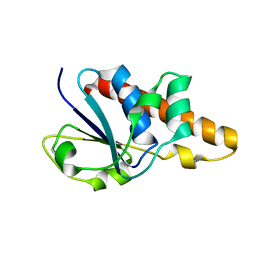 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
1Z2E
 
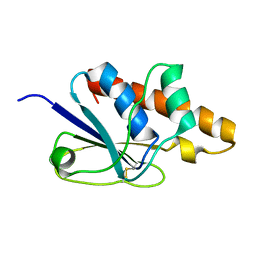 | | Solution Structure of Bacillus subtilis ArsC in oxidized state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
2MYP
 
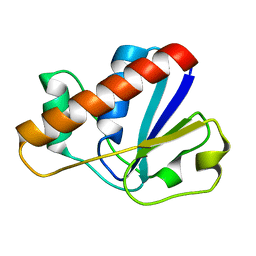 | |
2MYT
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYN
 
 | | An arsenate reductase in reduced state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Yu, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYU
 
 | | An arsenate reductase in oxidized state | | Descriptor: | Glutaredoxin arsenate reductase | | Authors: | Jin, C, Hu, C, Hu, Y. | | Deposit date: | 2015-01-30 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A Hybrid Mechanism for the Synechocystis Arsenate Reductase Revealed by Structural Snapshots during Arsenate Reduction.
J.Biol.Chem., 290, 2015
|
|
2MYJ
 
 | |
2M6S
 
 | | Holo_YqcA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2JTQ
 
 | | Rhodanese from E.coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
2JTR
 
 | | rhodanese persulfide from E. coli | | Descriptor: | Phage shock protein E | | Authors: | Jin, C, Li, H. | | Deposit date: | 2007-08-06 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of Escherichia coli rhodanese PspE in its sulfur-free and persulfide-intermediate forms: implications for the catalytic mechanism of rhodanese.
Biochemistry, 47, 2008
|
|
2M6R
 
 | | apo_YqcA | | Descriptor: | Flavodoxin | | Authors: | Jin, C, Hu, Y, Ye, Q. | | Deposit date: | 2013-04-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of the apo and holo states of flavodoxin YqcA from Escherichia coli.
Biomol.Nmr Assign., 8, 2014
|
|
2JSZ
 
 | | Solution structure of Tpx in the reduced state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J, Yang, F. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JSO
 
 | | Antimicrobial resistance protein | | Descriptor: | Polymyxin resistance protein pmrD | | Authors: | Jin, C, Fu, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
