2AZM
 
 | | Crystal structure of the MDC1 brct repeat in complex with the histone tail of gamma-H2AX | | Descriptor: | GAMMA-H2AX HISTONE, Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Stucki, M, Mohammad, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2005-09-12 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks
Cell(Cambridge,Mass.), 123, 2005
|
|
1Z56
 
 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
3I0N
 
 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
3I0M
 
 | | Structure of the S. pombe Nbs1 FHA/BRCT-repeat domain | | Descriptor: | DNA repair and telomere maintenance protein nbs1, GLYCEROL | | Authors: | Clapperton, J.A, Lloyd, J, Chapman, J.R, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2009-06-25 | | Release date: | 2009-10-13 | | Last modified: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage
Cell(Cambridge,Mass.), 139, 2009
|
|
1IK9
 
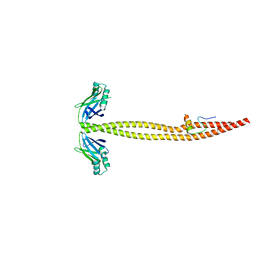 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
1GXC
 
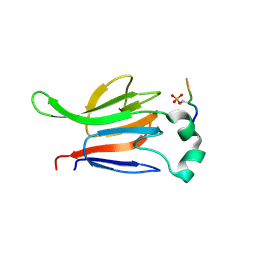 | | FHA domain from human Chk2 kinase in complex with a synthetic phosphopeptide | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE CHK2, SYNTHETIC PHOSPHOPEPTIDE | | Authors: | Li, J, Williams, B.L, Haire, L.F, Goldberg, M, Wilker, E, Durocher, D, Yaffe, M.B, Jackson, S.P, Smerdon, S.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-13 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Versatility of the Fha Domain in DNA-Damage Signaling by the Tumor Suppressor Kinase Chk2
Mol.Cell, 9, 2002
|
|
6HXV
 
 | |
4D2H
 
 | |
1H0Y
 
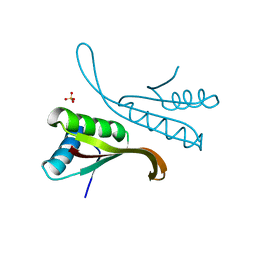 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B, SULFATE ION | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
2N5N
 
 | | Structure of an N-terminal domain of CHD4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Silva, A.P.G, Mackay, J.P. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The N-terminal Region of Chromodomain Helicase DNA-binding Protein 4 (CHD4) Is Essential for Activity and Contains a High Mobility Group (HMG) Box-like-domain That Can Bind Poly(ADP-ribose).
J.Biol.Chem., 291, 2016
|
|
1D3U
 
 | | TATA-BINDING PROTEIN/TRANSCRIPTION FACTOR (II)B/BRE+TATA-BOX COMPLEX FROM PYROCOCCUS WOESEI | | Descriptor: | DNA 23-MER: BRE+TATA-BOX, DNA 24-MER: BRE+TATA-BOX, TATA-BINDING PROTEIN, ... | | Authors: | Littlefield, O, Korkhin, Y, Sigler, P.B. | | Deposit date: | 1999-10-01 | | Release date: | 1999-11-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the oriented assembly of a TBP/TFB/promoter complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6HXT
 
 | |
6HXY
 
 | |
2QM4
 
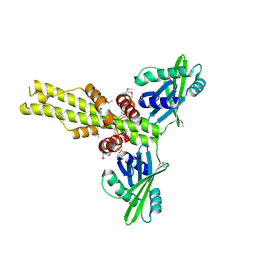 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
3WTD
 
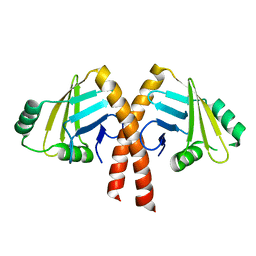 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
3WTF
 
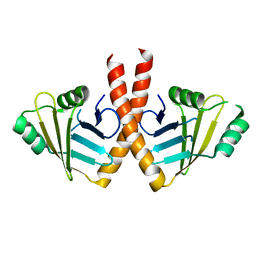 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1PCZ
 
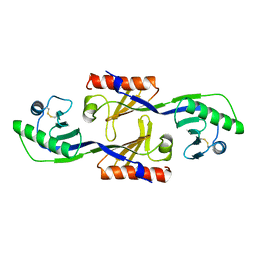 | | STRUCTURE OF TATA-BINDING PROTEIN | | Descriptor: | TATA-BINDING PROTEIN | | Authors: | Dedecker, B.S, Sigler, P.B. | | Deposit date: | 1996-10-04 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a hyperthermophilic archaeal TATA-box binding protein.
J.Mol.Biol., 264, 1996
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2N
 
 | |
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
4FCX
 
 | | S.pombe Mre11 apoenzym | | Descriptor: | DNA repair protein rad32, MANGANESE (II) ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FBW
 
 | |
4FBK
 
 | | Crystal structure of a covalently fused Nbs1-Mre11 complex with one manganese ion per active site | | Descriptor: | DNA repair and telomere maintenance protein nbs1,DNA repair protein rad32 CHIMERIC PROTEIN, MANGANESE (II) ION, SULFATE ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
