1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1A75
 
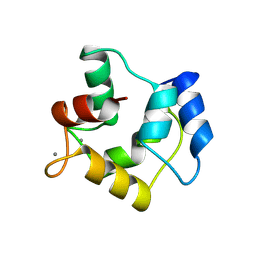 | | WHITING PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Baneres, J.L, Rambaud, J, Parello, J. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary Structure of a Trp-Containing Parvalbumin from Whiting (Merlangius Merlangus). Description of the Hydrophobic Core
To be Published
|
|
1BX7
 
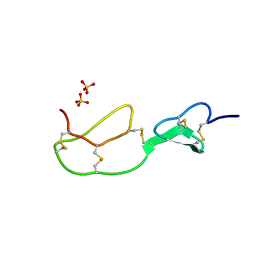 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.2 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BTL
 
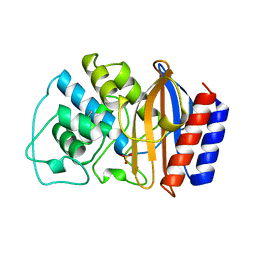 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
4RCR
 
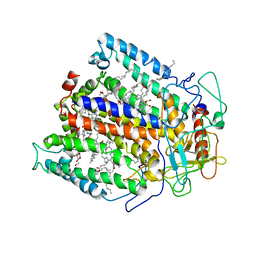 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
1BX8
 
 | | HIRUSTASIN FROM HIRUDO MEDICINALIS AT 1.4 ANGSTROMS | | Descriptor: | HIRUSTASIN, SULFATE ION | | Authors: | Uson, I, Sheldrick, G.M, De La Fortelle, E, Bricogne, G, Di Marco, S, Priestle, J.P, Gruetter, M.G, Mittl, P.R.E. | | Deposit date: | 1998-10-14 | | Release date: | 1999-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.2 A crystal structure of hirustasin reveals the intrinsic flexibility of a family of highly disulphide-bridged inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BMF
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Abrahams, J.P, Leslie, A.G.W, Lutter, R, Walker, J.E. | | Deposit date: | 1996-03-13 | | Release date: | 1996-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria.
Nature, 370, 1994
|
|
1BT5
 
 | | CRYSTAL STRUCTURE OF THE IMIPENEM INHIBITED TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PROTEIN (BETA-LACTAMASE), SULFATE ION | | Authors: | Maveyraud, L, Mourey, L, Pedelacq, J.D, Guillet, V, Kotra, L.K, Mobashery, S, Samama, J.P. | | Deposit date: | 1998-09-02 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A Beta-Lactamases from Antibiotic-Resistant Bacteria
J.Am.Chem.Soc., 120, 1998
|
|
1A62
 
 | | CRYSTAL STRUCTURE OF THE RNA-BINDING DOMAIN OF THE TRANSCRIPTIONAL TERMINATOR PROTEIN RHO | | Descriptor: | RHO | | Authors: | Allison, T.J, Wood, T.C, Briercheck, D.M, Rastinejad, F, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the RNA-binding domain from transcription termination factor rho.
Nat.Struct.Biol., 5, 1998
|
|
1A63
 
 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | Descriptor: | RHO | | Authors: | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
1ATT
 
 | |
1AY9
 
 | | WILD-TYPE UMUD' FROM E. COLI | | Descriptor: | UMUD PROTEIN | | Authors: | Peat, T.S, Frank, E.G, Mcdonald, J.P, Levine, A.S, Woodgate, R, Hendrickson, W.A. | | Deposit date: | 1997-11-15 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The UmuD' protein filament and its potential role in damage induced mutagenesis.
Structure, 4, 1996
|
|
1BY3
 
 | | FHUA FROM E. COLI | | Descriptor: | N-OCTYL-2-HYDROXYETHYL SULFOXIDE, PROTEIN (FERRICHROME-IRON RECEPTOR PRECURSOR (FHUA)) | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-22 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BY5
 
 | | FHUA FROM E. COLI, WITH ITS LIGAND FERRICHROME | | Descriptor: | FE (III) ION, FERRIC HYDROXAMATE UPTAKE PROTEIN, FERRICHROME, ... | | Authors: | Locher, K.P, Rees, B, Koebnik, R, Mitschler, A, Moulinier, L, Rosenbusch, J.P, Moras, D. | | Deposit date: | 1998-10-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes.
Cell(Cambridge,Mass.), 95, 1998
|
|
1CDT
 
 | | CARDIOTOXIN V4/II FROM NAJA MOSSAMBICA MOSSAMBICA: THE REFINED CRYSTAL STRUCTURE | | Descriptor: | CARDIOTOXIN VII4, PHOSPHATE ION | | Authors: | Rees, B, Bilwes, A, Samama, J.P, Moras, D. | | Deposit date: | 1990-05-17 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cardiotoxin VII4 from Naja mossambica mossambica. The refined crystal structure.
J.Mol.Biol., 214, 1990
|
|
9XIA
 
 | |
8XIA
 
 | |
1A71
 
 | | TERNARY COMPLEX OF AN ACTIVE SITE DOUBLE MUTANT OF HORSE LIVER ALCOHOL DEHYDROGENASE, PHE93=>TRP, VAL203=>ALA WITH NAD AND TRIFLUOROETHANOL | | Descriptor: | LIVER ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Bahnson, B.J, Chin, J.K, Klinman, J.P, Goldstein, B.M. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes.
Biochemistry, 37, 1998
|
|
1A72
 
 | | AN ACTIVE-SITE DOUBLE MUTANT (PHE93->TRP, VAL203->ALA) OF HORSE LIVER ALCOHOL DEHYDROGENASE IN COMPLEX WITH THE ISOSTERIC NAD ANALOG CPAD | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, HORSE LIVER ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Colby, T.D, Bahnson, B.J, Chin, J.K, Klinman, J.P, Goldstein, B.M. | | Deposit date: | 1998-03-19 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site modifications in a double mutant of liver alcohol dehydrogenase: structural studies of two enzyme-ligand complexes.
Biochemistry, 37, 1998
|
|
1AVB
 
 | | ARCELIN-1 FROM PHASEOLUS VULGARIS L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARCELIN-1, ... | | Authors: | Mourey, L, Pedelacq, J.D, Fabre, C, Rouge, P, Samama, J.P. | | Deposit date: | 1997-09-15 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the arcelin-1 dimer from Phaseolus vulgaris at 1.9-A resolution.
J.Biol.Chem., 273, 1998
|
|
1AP9
 
 | | X-RAY STRUCTURE OF BACTERIORHODOPSIN FROM MICROCRYSTALS GROWN IN LIPIDIC CUBIC PHASES | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Pebay-Peyroula, E, Rummel, G, Rosenbusch, J.P, Landau, E.M. | | Deposit date: | 1997-07-26 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray structure of bacteriorhodopsin at 2.5 angstroms from microcrystals grown in lipidic cubic phases.
Science, 277, 1997
|
|
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
5OCA
 
 | | PCSK9:Fab Complex with Dextran Sulfate | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-altropyranose-(1-6)-2,3-di-O-sulfo-alpha-L-glucopyranose, Fab from LDLR competitive antibody: Heavy chain, Fab from LDLR competitive antibody: Light chain, ... | | Authors: | Thirup, S.S, Vilstrup, J.P. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan sulfate proteoglycans present PCSK9 to the LDL receptor.
Nat Commun, 8, 2017
|
|
7UQT
 
 | | Solution NMR structure of hexahistidine tagged QseM (6H-QseM) | | Descriptor: | Quorum sensing master protein | | Authors: | Hall, D.A, Solomon, P.D, Bond, C.S, Ramsay, J.P, Mackay, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-20 | | Method: | SOLUTION NMR | | Cite: | DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria.
Nucleic Acids Res., 51, 2023
|
|
4V1S
 
 | | Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron | | Descriptor: | ALPHA-1,6-MANNANASE, GLYCEROL | | Authors: | Thompson, A.J, Cuskin, F, Spears, R.J, Dabin, J, Turkenburg, J.P, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2014-10-02 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Gh76 Alpha-Mannanase Homolog, Bt2949, from the Gut Symbiont Bacteroides Thetaiotaomicron
Acta Crystallogr.,Sect.D, 71, 2015
|
|
