2JBU
 
 | | Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CO-PURIFIED PEPTIDE, INSULIN-DEGRADING ENZYME | | Authors: | Im, H, Shen, Y, Tang, W.J. | | Deposit date: | 2006-12-11 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
3S46
 
 | | The crystal structure of alanine racemase from streptococcus pneumoniae | | Descriptor: | Alanine racemase, BENZOIC ACID | | Authors: | Im, H, Sharpe, M.L, Strych, U, Davlieva, M, Krause, K.L. | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of alanine racemase from Streptococcus pneumoniae, a target for structure-based drug design.
BMC MICROBIOL., 11, 2011
|
|
1IZ2
 
 | | Interactions causing the kinetic trap in serpin protein folding | | Descriptor: | alpha-D-glucopyranose-(1-2)-(5R)-5-[(2R)-2-hydroxynonyl]-beta-D-xylulofuranose, alpha1-antitrypsin | | Authors: | Im, H, Woo, M.-S, Hwang, K.Y, Yu, M.-H. | | Deposit date: | 2002-09-19 | | Release date: | 2003-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions causing the kinetic trap in serpin protein folding
J.BIOL.CHEM., 277, 2002
|
|
4NRN
 
 | | Crystal structure of metal-bound toxin from Helicobacter pylori | | Descriptor: | ZINC ION, metal-bound toxin | | Authors: | Lee, B.J, Im, H, Pathak, C, Jang, S.B. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of toxin HP0892 from Helicobacter pylori with two Zn(II) at 1.8 angstrom resolution
Protein Sci., 23, 2014
|
|
1XFC
 
 | | The 1.9 A crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | LeMagueres, P, Im, H, Ebalunode, J, Strych, U, Benedik, M.J, Briggs, J.M, Kohn, H, Krause, K.L. | | Deposit date: | 2004-09-14 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site.
Biochemistry, 44, 2005
|
|
1RCQ
 
 | | The 1.45 A crystal structure of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms | | Descriptor: | D-LYSINE, PYRIDOXAL-5'-PHOSPHATE, catabolic alanine racemase DadX | | Authors: | Le Magueres, P, Im, H, Dvorak, A, Strych, U, Benedik, M, Krause, K.L. | | Deposit date: | 2003-11-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45 A resolution of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms.
Biochemistry, 42, 2003
|
|
4LTT
 
 | | Crystal structure of native apo toxin from Helicobacter pylori | | Descriptor: | Uncharacterized protein, toxin | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4LS4
 
 | | Crystal structure of L66S mutant toxin from Helicobacter pylori | | Descriptor: | BROMIDE ION, Uncharacterized protein, Toxin | | Authors: | Pathak, C.C, Im, H, Lee, B.J, Yoon, H.J. | | Deposit date: | 2013-07-22 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4LSY
 
 | | Crystal structure of copper-bound L66S mutant toxin from Helicobacter pylori | | Descriptor: | CITRATE ANION, COPPER (II) ION, Uncharacterized protein, ... | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4Z1A
 
 | | Structure of apo form KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1D
 
 | | Structure of PEP and zinc bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHOENOLPYRUVATE, ZINC ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1B
 
 | | Structure of H204A mutant KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4Z1C
 
 | | Structure of Cadmium bound KDO8PS from H.pylori | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION | | Authors: | Lee, B.J, Cho, S, Im, H, Yoon, H.J. | | Deposit date: | 2015-03-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur.J.Med.Chem., 108, 2016
|
|
4N9H
 
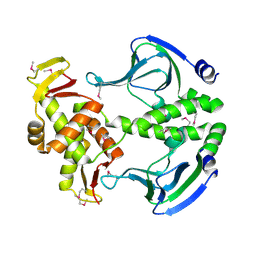 | | Crystal structure of Transcription regulation Protein CRP | | Descriptor: | Catabolite gene activator | | Authors: | Lee, B.J, Seok, S.H, Im, H, Yoon, H.J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N9I
 
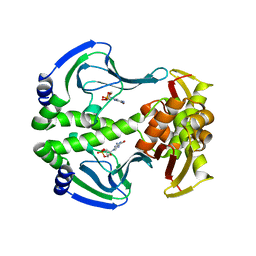 | | Crystal Structure of Transcription regulation protein CRP complexed with cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Catabolite gene activator | | Authors: | Lee, B.-J, Seok, S.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2013-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OF1
 
 | |
3CNK
 
 | | Crystal Structure of the dimerization domain of human filamin A | | Descriptor: | Filamin-A, SULFATE ION | | Authors: | Lee, B.J, Seo, M.D, Seok, S.H, Lee, S.J, Kwon, A.R, Im, H. | | Deposit date: | 2008-03-26 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the dimerization domain of human filamin A
Proteins, 75, 2008
|
|
4QJN
 
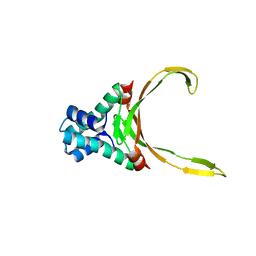 | | Crystal structure of apo nucleoid associated protein, SAV1473 | | Descriptor: | DNA-binding protein HU | | Authors: | Lee, B.-J, Kim, D.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | beta-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QJU
 
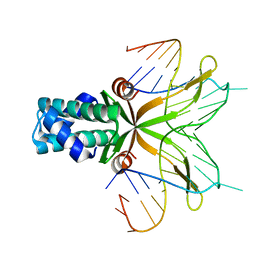 | | Crystal structure of DNA-bound nucleoid associated protein, SAV1473 | | Descriptor: | DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*CP*C)-3'), DNA-binding protein HU | | Authors: | Lee, B.-J, Kim, D.-H, Im, H, Yoon, H.-J. | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | beta-Arm flexibility of HU from Staphylococcus aureus dictates the DNA-binding and recognition mechanism
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2JG4
 
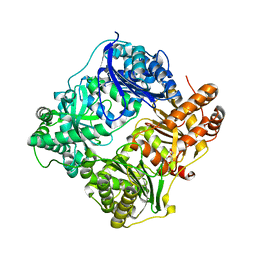 | | Substrate-free IDE structure in its closed conformation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
5NNV
 
 | |
7VHV
 
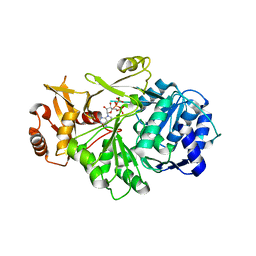 | | Crystal structure of S. aureus D-alanine alanyl carrier protein ligase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanyl carrier protein ligase, MAGNESIUM ION | | Authors: | Lee, B.J, Lee, I.-G, Im, H.G, Yoon, H.J. | | Deposit date: | 2021-09-23 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of the D-alanyl carrier protein ligase DltA from Staphylococcus aureus Mu50.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5NMO
 
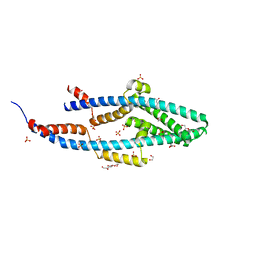 | | Structure of the Bacillus subtilis Smc Joint domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Chromosome partition protein Smc,Chromosome partition protein Smc, ... | | Authors: | Diebold-Durand, M.-L, Basquin, J, Gruber, S. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization.
Mol. Cell, 67, 2017
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
3NIP
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase complexed with 1,6-diaminohexane | | Descriptor: | 3-guanidinopropionase, HEXANE-1,6-DIAMINE | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
