6YM0
 
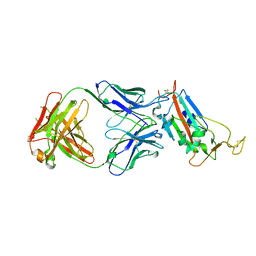 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YLA
 
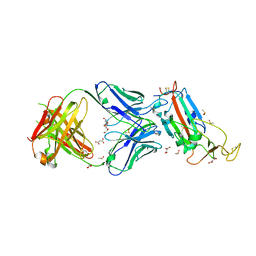 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
8BH5
 
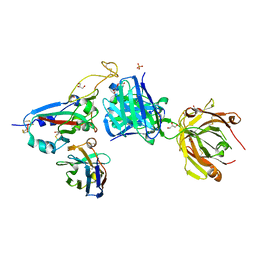 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
6Z97
 
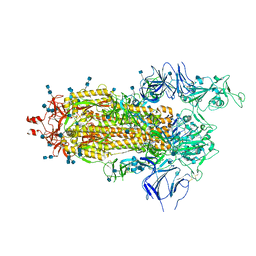 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
7ZXU
 
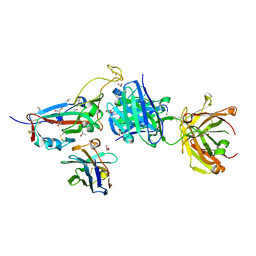 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
8ASY
 
 | | SARS-CoV-2 Omicron BA.2.75 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-08-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75.
Cell Rep, 42, 2022
|
|
7ZFF
 
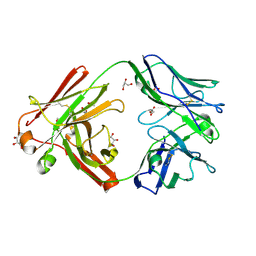 | | Omi-42 Fab | | Descriptor: | GLYCEROL, Omi-42 Heavy chain, Omi-42 light chain | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF6
 
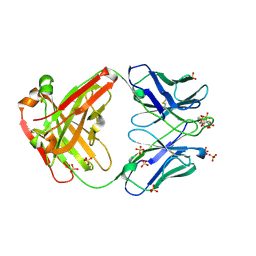 | | Omi-12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF8
 
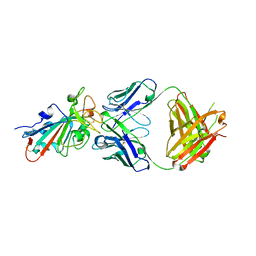 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFE
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-32 Fab and nanobody C1 | | Descriptor: | Nanobody C1, Omi-32 heavy chain, Omi-32 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFD
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-25 Fab | | Descriptor: | Omi-25 heavy chain, Omi-25 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF3
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-3 and EY6A Fabs | | Descriptor: | EY6A heavy chain, EY6A light chain, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFB
 
 | | SARS-CoV-2 Omicron RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody C1, Omi-18 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF9
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab (P21) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 heavy chain, COVOX-150 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF7
 
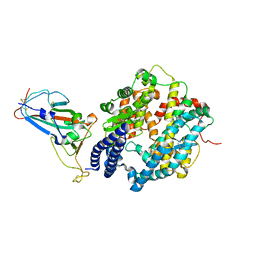 | | SARS-CoV-2 Omicron BA.2 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF5
 
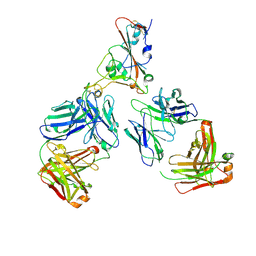 | | SARS-CoV-2 Omicron RBD in complex with Omi-12 and Beta-54 Fabs | | Descriptor: | Beta-54 heavy chain, Beta-54 light chain, Omi-12 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF4
 
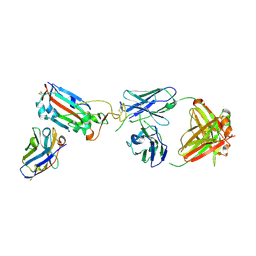 | | SARS-CoV-2 Omicron RBD in complex with Omi-9 Fab and nanobody F2 | | Descriptor: | Nanobody F2, Omi-9 heavy chain, Omi-9 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZFA
 
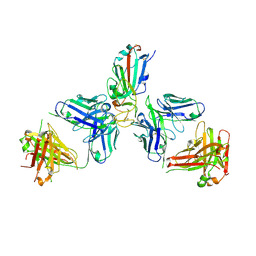 | | SARS-CoV-2 Omicron RBD in complex with Omi-6 and COVOX-150 Fabs | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Omi-6 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZAV
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA1
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | Authors: | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
