4JC2
 
 | | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral | | Descriptor: | asFP504 | | Authors: | Huang, Y.C, Emralino, F.L, Liu, F.C, Saloma, C.P, Bascos, N.A, Chen, C.J. | | Deposit date: | 2013-02-21 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Isolation, Cloning and Biophysical Analysis of a Novel Hexameric Green Fluorescent Protein from a Philippine Soft Coral
To be Published
|
|
6AHW
 
 | |
9IKZ
 
 | | SARS-CoV-2 E-RTC bound to pRNA-nsp9 and GDP-BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Helicase nsp13, ... | | Authors: | Yan, L.M, Huang, Y.C, Liu, Y.X, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2024-06-29 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Substrate selection and transition reveal the mechanism for RNA capping catalyzed by SARS-CoV-2 NiRAN
To Be Published
|
|
5XSK
 
 | | Crystal structure of PWWP-DNA complex for human hepatoma-derived growth factor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*TP*GP*GP*TP*CP*TP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*AP*GP*AP*CP*CP*A)-3'), ... | | Authors: | Chen, L.Y, Huang, Y.C, Hsieh, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2017-06-14 | | Release date: | 2018-06-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of PWWP-DNA complex at 2.84 Angstroms resolution
To Be Published
|
|
5XSL
 
 | |
2QN4
 
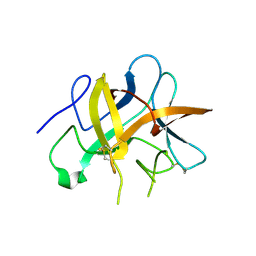 | | Structure and function study of rice bifunctional alpha-amylase/subtilisin inhibitor from Oryza sativa | | Descriptor: | Alpha-amylase/subtilisin inhibitor | | Authors: | Peng, W.Y, Lin, Y.H, Huang, Y.C, Guan, H.H, Hsieh, Y.C, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function Study of Rice Bifunctional Alpha-Amylase/Subtilisin Inhibitor from Oryza Sativa
To be Published
|
|
1Z5F
 
 | | Solution Structure of the Cytotoxic RC-RNase 3 with a Pyroglutamate Residue at the N-terminus | | Descriptor: | RC-RNase 3 | | Authors: | Lou, Y.C, Huang, Y.C, Pan, Y.R, Chen, C, Liao, Y.D. | | Deposit date: | 2005-03-18 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Roles of N-terminal pyroglutamate in maintaining structural integrity and pKa values of catalytic histidine residues in bullfrog ribonuclease 3
J.Mol.Biol., 355, 2006
|
|
3UZB
 
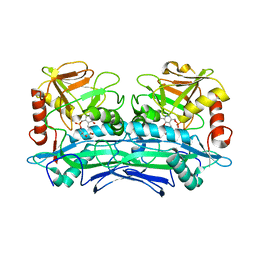 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UYY
 
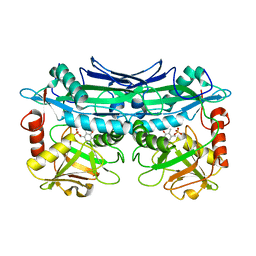 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZO
 
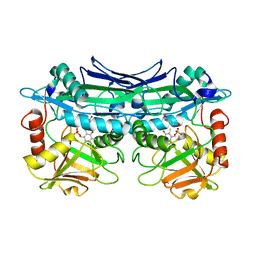 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
7EXF
 
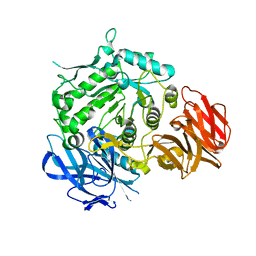 | | Crystal structure of wild-type from Arabidopsis thaliana complexed with Galactose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXJ
 
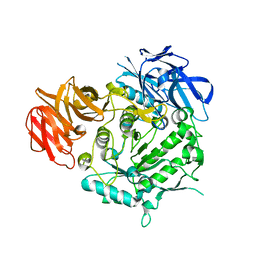 | | Crystal structure of alkaline alpha-galctosidase D383A mutant from Arabidopsis thaliana complexed with Raffinose | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXG
 
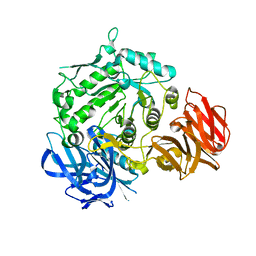 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXQ
 
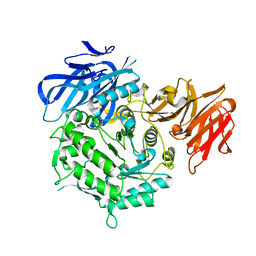 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with product-galactose and sucrose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, beta-D-galactopyranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXH
 
 | | Crystal structure of D383A mutant from Arabidopsis thaliana complexed with Galactinol. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, galactinol | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-27 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7EXR
 
 | | Crystal structure of alkaline alpha-galactosidase D383A mutant from Arabidopsis thaliana complexed with Stachyose. | | Descriptor: | Probable galactinol--sucrose galactosyltransferase 6, alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Chuankhayan, P, Guan, H.H, Lin, C.C, Chen, N.C, Huang, Y.C, Yoshimura, M, Nakagawa, A, Lee, R.H, Chen, C.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the hydrolase and synthase activities of an alkaline alpha-galactosidase from Arabidopsis from complexes with substrate/product.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5H0O
 
 | | Crystal structure of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease with manganese ion | | Descriptor: | HNH endonuclease, MANGANESE (II) ION | | Authors: | Zhang, L.K, Xu, D.D, Huang, Y.C, Gong, Y. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease.
Sci Rep, 7, 2017
|
|
5H0M
 
 | | Crystal structure of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease with zinc ion | | Descriptor: | HNH endonuclease, ZINC ION | | Authors: | Zhang, L.K, Xu, D.D, Huang, Y.C, Gong, Y. | | Deposit date: | 2016-10-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease
Sci Rep, 7, 2017
|
|
7EGQ
 
 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
7EIZ
 
 | | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Cell, 184, 2021
|
|
7BQ7
 
 | | Crystal structure of 2019-nCoV nsp16-nsp10 complex | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, S-ADENOSYLMETHIONINE, ... | | Authors: | Yan, L.M, Huang, Y.C, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of 2019-nCoV nsp16-nsp10 complex
To Be Published
|
|
7C1I
 
 | | Crystal structure of histidine-containing phosphotransfer protein B (HptB) from Pseudomonas aeruginosa PAO1 | | Descriptor: | Histidine kinase | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
7C1J
 
 | | Crystal structure of the receiver domain of sensor histidine kinase PA1611 (PA1611REC) from Pseudomonas aeruginosa PAO1 with magnesium ion coordinated in the active site cleft | | Descriptor: | Histidine kinase, MAGNESIUM ION | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
7CFW
 
 | | Crystal structure of the receiver domain of sensor histidine kinase PA1611 (PA1611REC) from Pseudomonas aeruginosa PAO1 with calcium ion coordinated in the active site cleft | | Descriptor: | CALCIUM ION, Histidine kinase | | Authors: | Chen, S.K, Guan, H.H, Wu, P.H, Lin, L.T, Wu, M.C, Chang, H.Y, Chen, N.C, Lin, C.C, Chuankhayan, P, Huang, Y.C, Lin, P.J, Chen, C.J. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural insights into the histidine-containing phospho-transfer protein and receiver domain of sensor histidine kinase suggest a complex model in the two-component regulatory system in Pseudomonas aeruginosa
Iucrj, 7, 2020
|
|
7YAT
 
 | | CryoEM tetra protofilament structure of the hamster prion 108-144 fibril | | Descriptor: | Major prion protein | | Authors: | Chen, E.H.-L, Kao, S.-W, Lee, C.-H, Huang, J.Y.C, Chen, R.P.-Y, Wu, K.-P. | | Deposit date: | 2022-06-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
J.Am.Chem.Soc., 144, 2022
|
|
