1Q6E
 
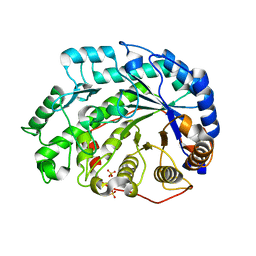 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 5.4 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6F
 
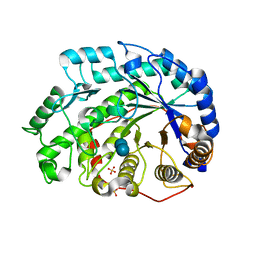 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 7.1 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6G
 
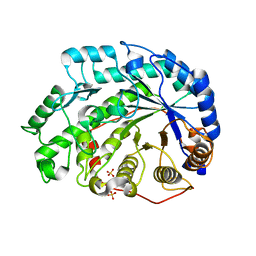 | | Crystal Structure of Soybean Beta-Amylase Mutant (N340T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
4FZ2
 
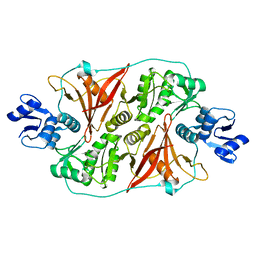 | | Crystal structure of the fourth type of archaeal tRNA splicing endonuclease from Candidatus Micrarchaeum acidiphilum ARMAN-2 | | Descriptor: | tRNA intron endonuclease | | Authors: | Hirata, A, Fujishima, K, Yamagami, R, Kawamura, T, Banfiled, J.F, Kanai, A, Hori, H. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity
Nucleic Acids Res., 40, 2012
|
|
5E71
 
 | |
5E72
 
 | |
3P1Z
 
 | | Crystal structure of the Aperopyrum pernix RNA splicing endonuclease | | Descriptor: | Putative uncharacterized protein, tRNA-splicing endonuclease | | Authors: | Hirata, A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop
Nucleic Acids Res., 39, 2011
|
|
3P1Y
 
 | |
2PA8
 
 | |
1VEM
 
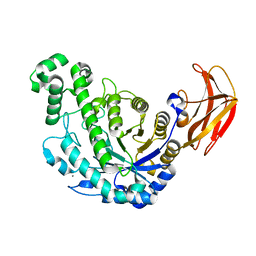 | | Crystal Structure Analysis of Bacillus Cereus Beta-Amylase at the optimum pH (6.5) | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEN
 
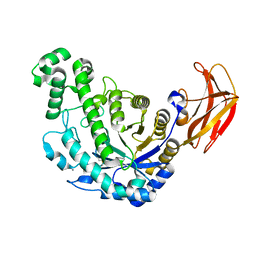 | | Crystal Structure Analysis of Y164E/maltose of Bacilus cereus Beta-amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEO
 
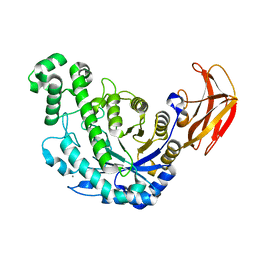 | | Crystal Structure Analysis of Y164F/maltose of Bacillus cereus Beta-Amylase at pH 4.6 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1VEP
 
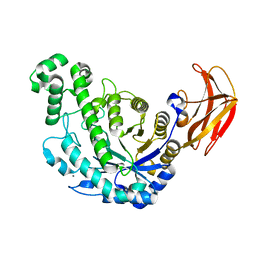 | | Crystal Structure Analysis of Triple (T47M/Y164E/T328N)/maltose of Bacillus cereus Beta-Amylase at pH 6.5 | | Descriptor: | Beta-amylase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-04-03 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Engineering of the pH optimum of Bacillus cereus beta-amylase: conversion of the pH optimum from a bacterial type to a higher-plant type
Biochemistry, 43, 2004
|
|
1Q6D
 
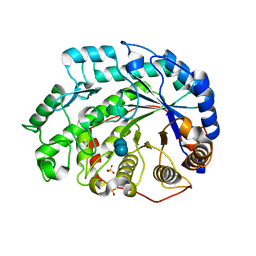 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6C
 
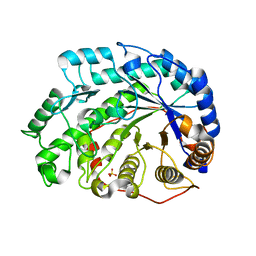 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
6JPL
 
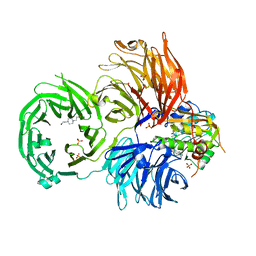 | | The X-ray structure of yeast tRNA methyltransferase Trm7-Trm734 in complex with S-adenosyl-L-methionine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Shimizu, N, Hori, H. | | Deposit date: | 2019-03-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6JP6
 
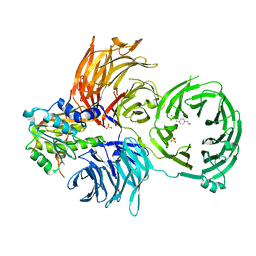 | | The X-ray structure of yeast tRNA methyltransferase complex of Trm7 and Trm734 essential for 2'-O-methylation at the first position of anticodon in specific tRNAs | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, tRNA (cytidine(34)/guanosine(34)-2'-O)-methyltransferase, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Sihimzu, N, Hori, H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
4QJV
 
 | |
7EDC
 
 | | Crystal structure of mutant tRNA [Gm18] methyltransferase TrmH (E107G) in complex with S-adenosyl-L-methionine from Escherichia coli | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanosine(18)-2'-O)-methyltransferase | | Authors: | Kono, Y, Ito, A, Okamoto, A, Yamagami, R, Hirata, A, Hori, H. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Unique substrate specificity of type II tRNA Gm18 methyltransferase from Escherichia coli
To Be Published
|
|
5X89
 
 | | The X-ray crystal structure of subunit fusion RNA splicing endonuclease from Methanopyrus kandleri | | Descriptor: | EndA-like protein,tRNA-splicing endonuclease, PHOSPHATE ION | | Authors: | Kaneta, A, Fujishima, K, Morikazu, W, Hori, H, Hirata, A. | | Deposit date: | 2017-03-01 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The RNA-splicing endonuclease from the euryarchaeaon Methanopyrus kandleri is a heterotetramer with constrained substrate specificity
Nucleic Acids Res., 46, 2018
|
|
1CQY
 
 | | STARCH BINDING DOMAIN OF BACILLUS CEREUS BETA-AMYLASE | | Descriptor: | BETA-AMYLASE | | Authors: | Yoon, H.J, Hirata, A, Adachi, M, Sekine, A, Utsumi, S, Mikami, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Separated Starch-Binding Domain of Bacillus cereus B-amylase
To be Published
|
|
8JL8
 
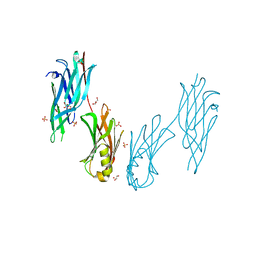 | | Crystal structure of the collagen binding domain of Cnm from Streptococcus mutans | | Descriptor: | Collagen-binding adhesin, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S.-i, Hirata, A, Takano, K. | | Deposit date: | 2023-06-02 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure, Stability and Binding Properties of Collagen-Binding Domains from Streptococcus mutans.
Chemistry, 5, 2023
|
|
3W5B
 
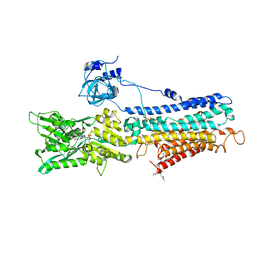 | | Crystal structure of the recombinant SERCA1a (calcium pump of fast twitch skeletal muscle) in the E1.Mg2+ state | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Toyoshima, C, Iwasawa, S, Ogawa, H, Hirata, A, Tsueda, J, Inesi, G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state.
Nature, 495, 2013
|
|
3W5C
 
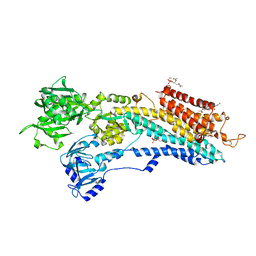 | | Crystal structure of the calcium pump in the E2 state free from exogenous inhibitors | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, SERCA1a, SODIUM ION | | Authors: | Toyoshima, C, Iwasawa, S, Ogawa, H, Hirata, A, Tsueda, J, Inesi, G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state.
Nature, 495, 2013
|
|
3W5A
 
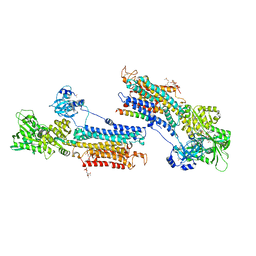 | | Crystal structure of the calcium pump and sarcolipin from rabbit fast twitch skeletal muscle in the E1.Mg2+ state | | Descriptor: | 2',3'-O-[(1r)-2,4,6-trinitrocyclohexa-2,5-diene-1,1-diyl]adenosine 5'-(dihydrogen phosphate), MAGNESIUM ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Toyoshima, C, Iwasawa, S, Ogawa, H, Hirata, A, Tsueda, J, Inesi, G. | | Deposit date: | 2013-01-27 | | Release date: | 2013-03-06 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of the calcium pump and sarcolipin in the Mg2+-bound E1 state.
Nature, 495, 2013
|
|
