5WWP
 
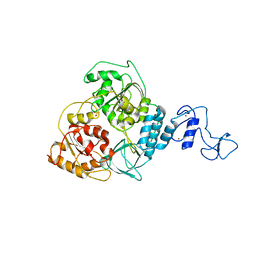 | | Crystal structure of Middle East respiratory syndrome coronavirus helicase (MERS-CoV nsp13) | | Descriptor: | ORF1ab, SULFATE ION, ZINC ION | | Authors: | Hao, W, Wojdyla, J.A, Zhao, R, Han, R, Das, R, Zlatev, I, Manoharan, M, Wang, M, Cui, S. | | Deposit date: | 2017-01-03 | | Release date: | 2017-07-05 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Middle East respiratory syndrome coronavirus helicase
PLoS Pathog., 13, 2017
|
|
1HHV
 
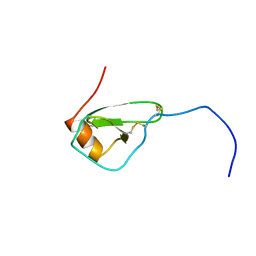 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
1MI2
 
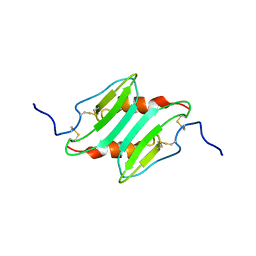 | | SOLUTION STRUCTURE OF MURINE MACROPHAGE INFLAMMATORY PROTEIN-2, NMR, 20 STRUCTURES | | Descriptor: | MACROPHAGE INFLAMMATORY PROTEIN-2 | | Authors: | Shao, W, Jerva, L.F, West, J, Lolis, E, Schweitzer, B.I. | | Deposit date: | 1997-10-24 | | Release date: | 1998-04-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of murine macrophage inflammatory protein-2.
Biochemistry, 37, 1998
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XK8
 
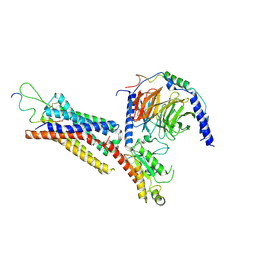 | | Cryo-EM structure of the Neuromedin U receptor 2 (NMUR2) in complex with G Protein and its endogeneous Peptide-Agonist NMU25 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, W, Wenru, Z, Mu, W, Minmin, L, Shutian, C, Tingting, T, Gisela, S, Holger, W, Albert, B, Cuiying, Y, Xiaojing, C, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand recognition and activation of neuromedin U receptor 2.
Nat Commun, 13, 2022
|
|
4M70
 
 | |
5JCB
 
 | | Microtubule depolymerizing agent podophyllotoxin derivative YJTSF1 | | Descriptor: | (5R,5aR,8aS,9R)-9-[(4H-1,2,4-triazol-3-yl)sulfanyl]-5-(3,4,5-trimethoxyphenyl)-5,8,8a,9-tetrahydro-2H-furo[3',4':6,7]naphtho[2,3-d][1,3]dioxol-6(5aH)-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Guan, Z, Zhao, W, Yin, P. | | Deposit date: | 2016-04-14 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4 beta-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin.
ACS Chem. Biol., 12, 2017
|
|
7FEX
 
 | |
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
5X41
 
 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
6PKM
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#8) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.17 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKQ
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.85A resolution (#11) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
6IMA
 
 | | Crystal Structure of ALKBH1 without alpha-1 (N37-C369) | | Descriptor: | CITRIC ACID, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6PKS
 
 | | MicroED structure of proteinase K from low-dose merged lamellae that were not pre-coated with platinum 2.16A resolution (LD) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.16 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKL
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.59A resolution (#7) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.59 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKT
 
 | | MicroED structure of proteinase K from merging low-dose, platinum pre-coated lamellae at 1.85A resolution (LDPT) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKN
 
 | | MicroED structure of proteinase K from an unpolished, platinum-coated, single lamella at 2.08A resolution (#9) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.08 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKJ
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#2) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKO
 
 | | MicroED structure of proteinase K from a platinum coated, unpolished, single lamella at 2.07A resolution (#12) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.07 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKR
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.79A resolution (#13) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.79 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKK
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.18A resolution (#5) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
