1M6F
 
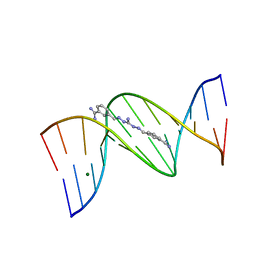 | | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove | | Descriptor: | 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Nguyen, B, Lee, M.P.H, Hamelberg, D, Joubert, A, Bailly, C, Brun, R, Neidle, S, Wilson, W.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
J.Am.Chem.Soc., 124, 2002
|
|
3FSM
 
 | |
3NWQ
 
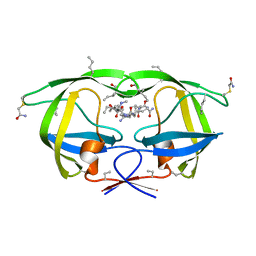 | |
6PLD
 
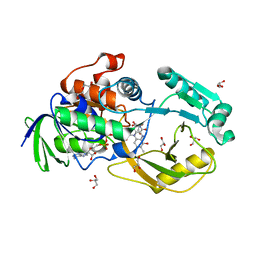 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6P9D
 
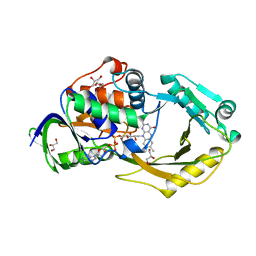 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
9BSH
 
 | |
9BSM
 
 | |
3NXE
 
 | |
3NXN
 
 | | X-ray structure of ester chemical analogue 'covalent dimer' [Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease covalent dimer | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-14 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NWX
 
 | | X-ray structure of ester chemical analogue [O-Ile50,O-Ile50']HIV-1 protease complexed with KVS-1 inhibitor | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, SULFATE ION, protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2010-07-12 | | Release date: | 2011-11-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NYG
 
 | |
4HSJ
 
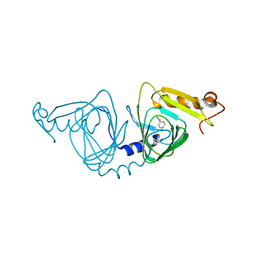 | | 1.88 angstrom x-ray crystal structure of piconlinic-bound 3-hydroxyanthranilate-3,4-dioxygenase | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION, PYRIDINE-2-CARBOXYLIC ACID | | Authors: | Liu, F, Chen, L, Davis, C.I, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
4HVO
 
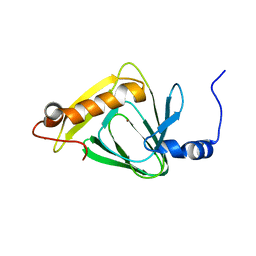 | | 1.75 angstrom x-ray crystal structure of cufe reconstituted 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, COPPER (II) ION, FE (II) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
4HVQ
 
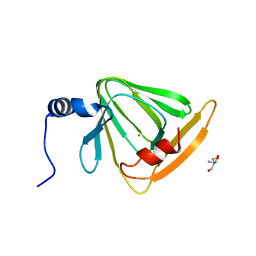 | | X-ray crystal structure of FECU reconstituted 3-hydroxyanthranilate-3,4-dioxygenase from cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
4L2N
 
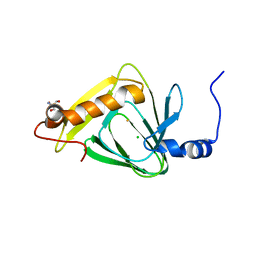 | | Understanding Extradiol Dioxygenase Mechanism in NAD+ Biosynthesis by Viewing Catalytic Intermediates - ligand-free structure | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, CHLORIDE ION, ... | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2013-06-04 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
3HDK
 
 | | Crystal structure of chemically synthesized [Aib51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [Aib51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IAW
 
 | |
3HLO
 
 | |
3HBO
 
 | | Crystal structure of chemically synthesized [D-Ala51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [D-Ala51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3HAW
 
 | |
3HAU
 
 | |
3KA2
 
 | |
