4V4B
 
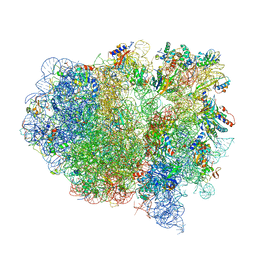 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
5WEN
 
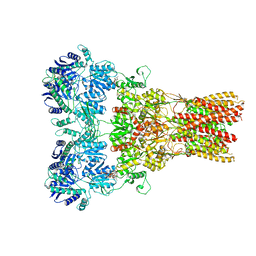 | | GluA2 bound to GSG1L in digitonin, state 2 | | Descriptor: | Digitonin, Glutamate receptor 2,Germ cell-specific gene 1-like protein | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
4V69
 
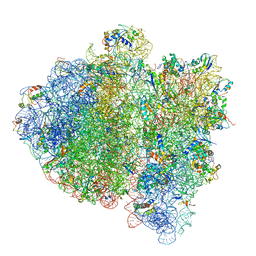 | | Ternary complex-bound E.coli 70S ribosome. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Villa, E, Sengupta, J, Trabuco, L.G, LeBarron, J, Baxter, W.T, Shaikh, T.R, Grassucci, R.A, Nissen, P, Ehrenberg, M, Schulten, K, Frank, J. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V8M
 
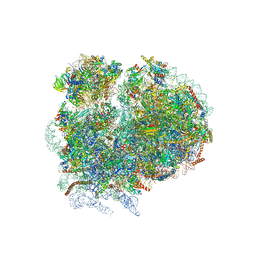 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
4C4Q
 
 | | Cryo-EM map of the CSFV IRES in complex with the small ribosomal 40S subunit and DHX29 | | Descriptor: | INTERNAL RIBOSOMAL ENTRY SITE | | Authors: | Hashem, Y, desGeorges, A, Dhote, V, Langlois, R, Liao, H.Y, Grassucci, R.A, Pestova, T.V, Hellen, C.U.T, Frank, J. | | Deposit date: | 2013-09-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Hepatitis-C-Virus-Like Internal Ribosome Entry Sites Displace Eif3 to Gain Access to the 40S Subunit
Nature, 503, 2013
|
|
5T5H
 
 | | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit | | Descriptor: | 40S ribosomal protein L14, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liu, Z, Gutierrez-Vargas, C, Wei, J, Grassucci, R.A, Ramesh, M, Espina, N, Sun, M, Tutuncuoglu, B, Madison-Antenucci, S, Woolford Jr, J.L, Tong, L, Frank, J. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6BO8
 
 | | Cryo-EM structure of human TRPV6 in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3J5Y
 
 | | Structure of the mammalian ribosomal pre-termination complex associated with eRF1-eRF3-GDPNP | | Descriptor: | 5'-R(*AP*UP*UP*GP*UP*AP*AP*AP*AP*A)-3', Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1, ... | | Authors: | des Georges, A, Hashem, Y, Unbehaun, A, Grassucci, R.A, Taylor, D, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
Nucleic Acids Res., 42, 2014
|
|
1MVR
 
 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | Descriptor: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | Authors: | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1MI6
 
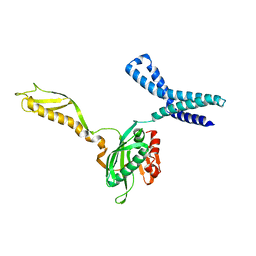 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
5KBS
 
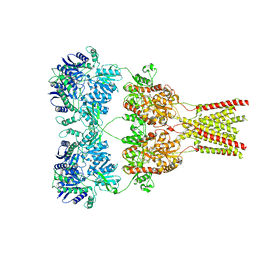 | | Cryo-EM structure of GluA2-0xSTZ at 8.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBU
 
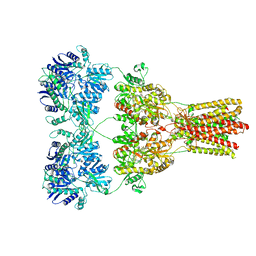 | | Cryo-EM structure of GluA2-2xSTZ complex at 7.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
5KBT
 
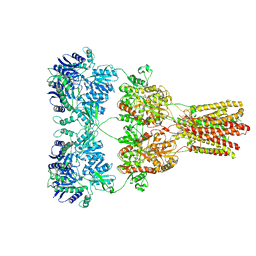 | | Cryo-EM structure of GluA2-1xSTZ complex at 6.4 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy.
Science, 353, 2016
|
|
1FCW
 
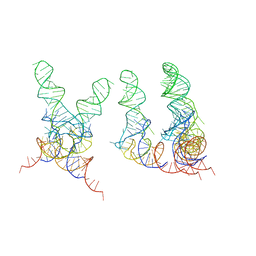 | | TRNA POSITIONS DURING THE ELONGATION CYCLE | | Descriptor: | TRNAPHE | | Authors: | Agrawal, R.K, Spahn, C.M.T, Penczek, P, Grassucci, R.A, Nierhaus, K.H, Frank, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-08-11 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle.
J.Cell Biol., 150, 2000
|
|
1RY1
 
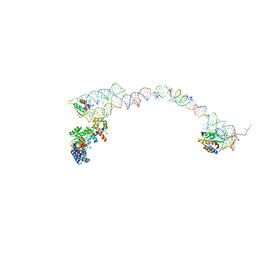 | | Structure of the signal recognition particle interacting with the elongation-arrested ribosome | | Descriptor: | SRP Alu domain, SRP RNA, SRP S domain, ... | | Authors: | Halic, M, Becker, T, Pool, M.R, Spahn, C.M, Grassucci, R.A, Frank, J, Beckmann, R. | | Deposit date: | 2003-12-19 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the signal recognition particle interacting with the elongation-arrested ribosome
Nature, 427, 2004
|
|
1T1O
 
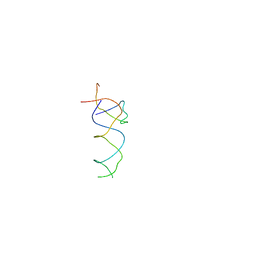 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | Descriptor: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
1T1M
 
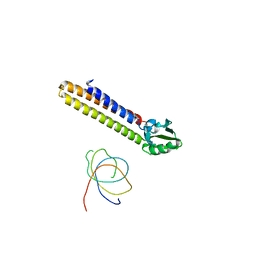 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6BO9
 
 | | Cryo-EM structure of human TRPV6 in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BOA
 
 | | Cryo-EM structure of human TRPV6-R470E in amphipols | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6BOB
 
 | | Cryo-EM structure of rat TRPV6* in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 6 | | Authors: | McGoldrick, L.L, Singh, A.K, Saotome, K, Yelshanskaya, M.V, Twomey, E.C, Grassucci, R.A, Sobolevsky, A.I. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Opening of the human epithelial calcium channel TRPV6.
Nature, 553, 2018
|
|
6OT3
 
 | | RF2 accommodated state bound Release complex 70S at 24 ms | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fu, Z, Indrisiunaite, G, Kaledhonkar, S, Shah, B, Sun, M, Chen, B, Grassucci, R.A, Ehrenberg, M, Frank, J. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The structural basis for release-factor activation during translation termination revealed by time-resolved cryogenic electron microscopy.
Nat Commun, 10, 2019
|
|
