3F7T
 
 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3KEF
 
 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEM
 
 | | Crystal structure of IspH:IPP complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE9
 
 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEL
 
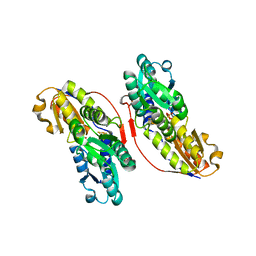 | | Crystal Structure of IspH:PP complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PYROPHOSPHATE 2- | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE8
 
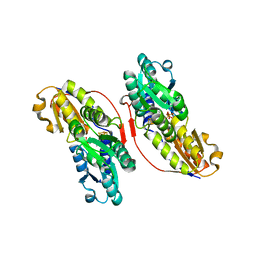 | | Crystal structure of IspH:HMBPP-complex | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4UQF
 
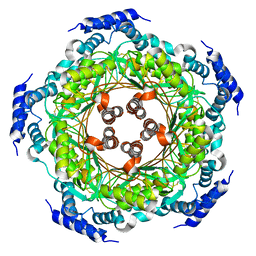 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES GTP CYCLOHYDROLASE I | | Descriptor: | GTP cyclohydrolase 1 | | Authors: | Schuessler, S, Perbandt, M, Fischer, M, Graewert, T. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of GTP cyclohydrolase I from Listeria monocytogenes, a potential anti-infective drug target.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3R0I
 
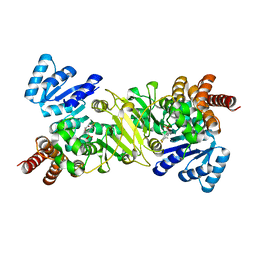 | | IspC in complex with an N-methyl-substituted hydroxamic acid | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, {(1S)-1-(3,4-difluorophenyl)-4-[hydroxy(methyl)amino]-4-oxobutyl}phosphonic acid | | Authors: | Behrendt, C.T, Kunfermann, A, Illarionova, V, Matheeussen, A, Pein, M.K, Graewert, T, Bacher, A, Eisenreich, W, Illarionov, B, Fischer, M, Maes, L, Groll, M, Kurz, T. | | Deposit date: | 2011-03-08 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reverse Fosmidomycin Derivatives against the Antimalarial Drug Target IspC (Dxr).
J.Med.Chem., 54, 2011
|
|
3NOY
 
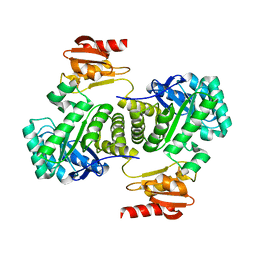 | | Crystal structure of IspG (gcpE) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Bacher, A. | | Deposit date: | 2010-06-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG.
J.Mol.Biol., 404, 2010
|
|
3SZL
 
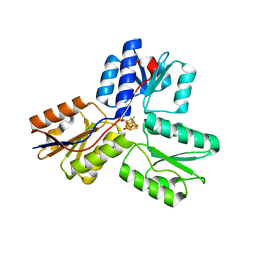 | | IspH:Ligand Mutants - wt 70sec | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3SZO
 
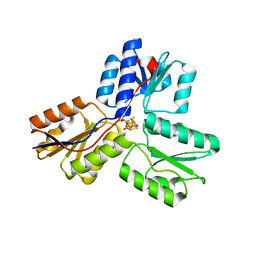 | | IspH:HMBPP complex after 3 minutes X-ray pre-exposure | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3SZU
 
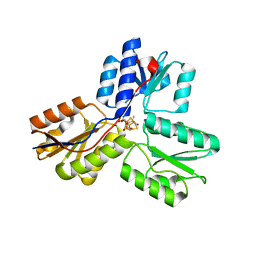 | | IspH:HMBPP complex structure of E126Q mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3T0G
 
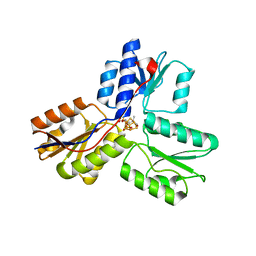 | | IspH:HMBPP (substrate) structure of the T167C mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3T0F
 
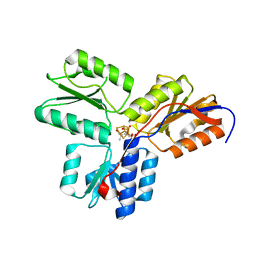 | | IspH:HMBPP (substrate) structure of the E126D mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
