1YUP
 
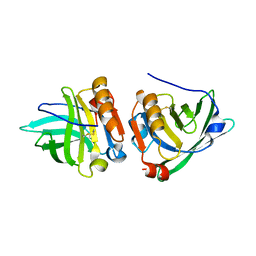 | | Reindeer beta-lactoglobulin | | Descriptor: | beta-lactoglobulin | | Authors: | Goldman, A, Oksanen, E. | | Deposit date: | 2005-02-14 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reindeer beta-lactoglobulin crystal structure with pseudo-body-centred noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8PRK
 
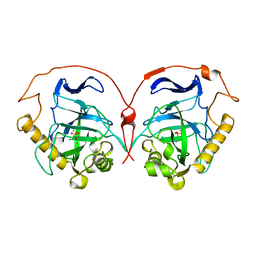 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
2V5E
 
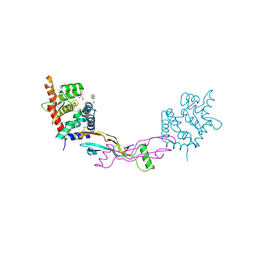 | | The structure of the GDNF:Coreceptor complex: Insights into RET signalling and heparin binding. | | Descriptor: | 1,2-ETHANEDIOL, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Parkash, V, Leppanen, V.-M, Virtanen, H, Jurvansuu, J.-M, Bespalov, M.M, Sidorova, Y.A, Runeberg-Roos, P, Saarma, M, Goldman, A. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structure of the Glial Cell Line-Derived Neurotrophic Factor-Coreceptor Complex: Insights Into Ret Signaling and Heparin Binding.
J.Biol.Chem., 283, 2008
|
|
7O23
 
 | |
1IPW
 
 | |
9G8K
 
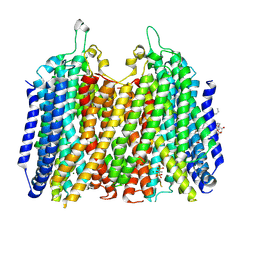 | | Structure of K+-dependent Na+-PPase from Thermotoga maritima in complex with Ca2+ and Etidronate | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Vidilaseris, K, Liu, J, Goldman, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformational dynamics and asymmetry in multimodal inhibition of membrane-bound pyrophosphatases
Elife, 2024
|
|
9G8J
 
 | |
3MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT I54V | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-27 | | Release date: | 1999-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
1MUC
 
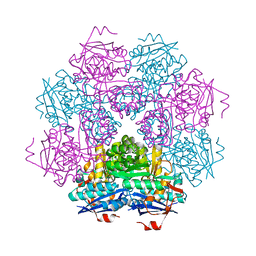 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
8CHA
 
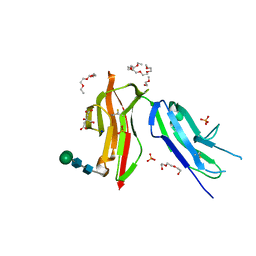 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
2IK7
 
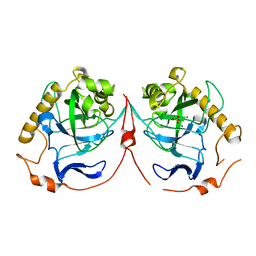 | | Yeast inorganic pyrophosphatase variant D120N with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IHP
 
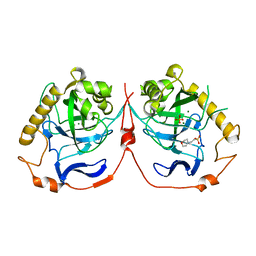 | | Yeast inorganic pyrophosphatase with magnesium and phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK0
 
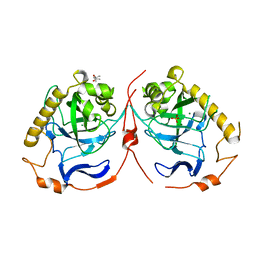 | | Yeast inorganic pyrophosphatase variant E48D with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
1QHM
 
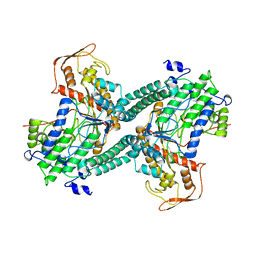 | | ESCHERICHIA COLI PYRUVATE FORMATE LYASE LARGE DOMAIN | | Descriptor: | PYRUVATE FORMATE-LYASE | | Authors: | Leppanen, V.-M, Merckel, M.C, Ollis, D.L, Wong, K.K, Kozarich, J.W, Goldman, A. | | Deposit date: | 1999-05-19 | | Release date: | 2000-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyruvate formate lyase is structurally homologous to type I ribonucleotide reductase.
Structure Fold.Des., 7, 1999
|
|
6QXA
 
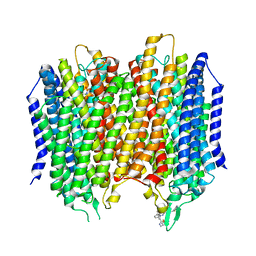 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
2MUC
 
 | | MUCONATE CYCLOISOMERASE VARIANT F329I | | Descriptor: | MANGANESE (II) ION, PROTEIN (MUCONATE CYCLOISOMERASE) | | Authors: | Schell, U, Helin, S, Kajander, T, Schlomann, M, Goldman, A. | | Deposit date: | 1998-10-26 | | Release date: | 1999-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the activity of two muconate cycloisomerase variants toward substituted muconates.
Proteins, 34, 1999
|
|
8B22
 
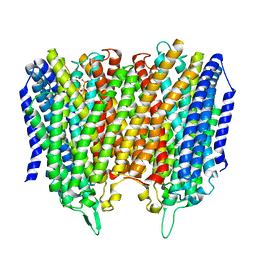 | |
8B23
 
 | |
8B21
 
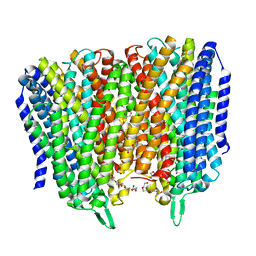 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 0-60-seconds post reaction initiation with Na+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-BETA-D-MALTOSIDE, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B37
 
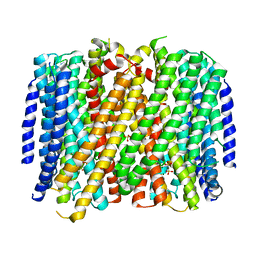 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
8B24
 
 | | Time-resolved structure of K+-dependent Na+-PPase from Thermotoga maritima 3600-seconds post reaction initiation with Na+ | | Descriptor: | DIPHOSPHATE, K(+)-stimulated pyrophosphate-energized sodium pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Vidilaseris, K, Goldman, A. | | Deposit date: | 2022-09-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
2EIP
 
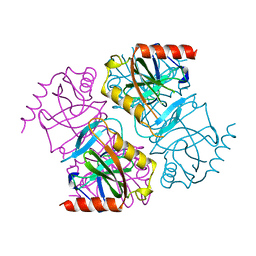 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1FAJ
 
 | | INORGANIC PYROPHOSPHATASE | | Descriptor: | SOLUBLE INORGANIC PYROPHOSPHATASE | | Authors: | Kankare, J.A, Salminen, T, Goldman, A. | | Deposit date: | 1996-01-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Escherichia coli inorganic pyrophosphatase at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1JOF
 
 | | Neurospora crassa 3-carboxy-cis,cis-mucoante lactonizing enzyme | | Descriptor: | BETA-MERCAPTOETHANOL, CARBOXY-CIS,CIS-MUCONATE CYCLASE, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Kajander, T, Merckel, M.C, Thompson, A, Deacon, A.M, Mazur, P, Kozarich, J.W, Goldman, A. | | Deposit date: | 2001-07-28 | | Release date: | 2002-04-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Neurospora crassa 3-carboxy-cis,cis-muconate lactonizing enzyme, a beta propeller cycloisomerase.
Structure, 10, 2002
|
|
6QP4
 
 | | Structure of 299-452 fragment of the UspA1 protein from Moraxella catarrhalis | | Descriptor: | CHLORIDE ION, HEXANE-1,6-DIOL, UspA1, ... | | Authors: | Mikula, K.M, Kolodziejczyk, R, Goldman, A. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the UspA1 protein fragment from Moraxella catarrhalis responsible for C3d binding.
J.Struct.Biol., 208, 2019
|
|
