1X8H
 
 | |
1X8I
 
 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
3G5I
 
 | | Crystal Structure of the E.coli RihA pyrimidine nucleosidase bound to a iminoribitol-based inhibitor | | Descriptor: | (2S,3S,4R,5R)-2-(3,4-diaminophenyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Garau, G, Muzzolini, L, Tornaghi, P, Degano, M. | | Deposit date: | 2009-02-05 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active site plasticity revealed from the structure of the enterobacterial N-ribohydrolase RihA bound to a competitive inhibitor.
Bmc Struct.Biol., 10, 2010
|
|
1X8G
 
 | |
1WRA
 
 | |
1HH7
 
 | |
2FPH
 
 | |
6FLL
 
 | |
1I8O
 
 | |
2QDS
 
 | |
1I8P
 
 | |
2QDT
 
 | |
7O2V
 
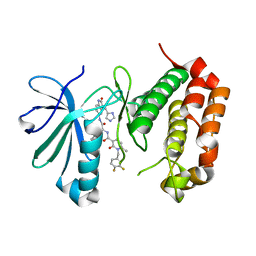 | | AURORA KINASE A IN COMPLEX WITH THE AUR-A/PDK1 INHIBITOR VI8 | | Descriptor: | 1-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N}-[(1~{R})-2-[[(3~{E})-3-(1~{H}-imidazol-5-ylmethylidene)-2-oxidanylidene-1~{H}-indol-5-yl]amino]-2-oxidanylidene-1-phenyl-ethyl]-6-methyl-2-oxidanylidene-pyridine-3-carboxamide, Aurora kinase A | | Authors: | Garau, G. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of potent dual PDK1/AurA kinase inhibitors for cancer therapy: Lead-optimization, structural insights, and ADME-Tox profile.
Eur.J.Med.Chem., 226, 2021
|
|
4QN9
 
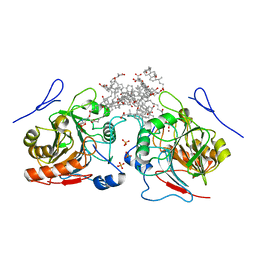 | | Structure of human NAPE-PLD | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Structure of human N-acylphosphatidylethanolamine-hydrolyzing phospholipase D: regulation of fatty acid ethanolamide biosynthesis by bile acids.
Structure, 23, 2015
|
|
4DO3
 
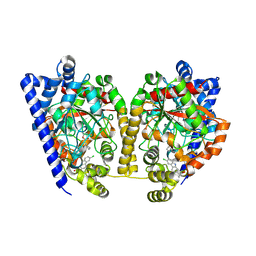 | | Structure of FAAH with a non-steroidal anti-inflammatory drug | | Descriptor: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CHLORIDE ION, CYCLOHEXANE AMINOCARBOXYLIC ACID, ... | | Authors: | Garau, G. | | Deposit date: | 2012-02-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Binding Site for Nonsteroidal Anti-inflammatory Drugs in Fatty Acid Amide Hydrolase.
J.Am.Chem.Soc., 135, 2013
|
|
8P96
 
 | | TARGET COMPLEX 3 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-26 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | NAPE-PLD is target of thiazide diuretics.
Cell Chem Biol, 32, 2025
|
|
8P90
 
 | | TARGET COMPLEX 2 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-26 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAPE-PLD is target of thiazide diuretics.
Cell Chem Biol, 32, 2025
|
|
8PC4
 
 | | MEMBRANE TARGET COMPLEX 1 | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, ... | | Authors: | Garau, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-26 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | NAPE-PLD is target of thiazide diuretics.
Cell Chem Biol, 32, 2025
|
|
2GKL
 
 | | Crystal structure of the zinc carbapenemase CPHA in complex with the inhibitor pyridine-2,4-dicarboxylate | | Descriptor: | Beta-lactamase, GLYCEROL, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2006-04-03 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Competitive Inhibitors of the CphA Metallo-{beta}-Lactamase from Aeromonas hydrophila
Antimicrob.Agents Chemother., 51, 2007
|
|
3MKN
 
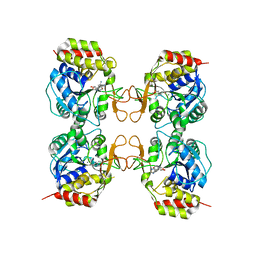 | | Crystal structure of the E. coli pyrimidine nucleosidase YeiK bound to a competitive inhibitor | | Descriptor: | (2S,3S,4R,5R)-2-(3,4-diaminophenyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
3MKM
 
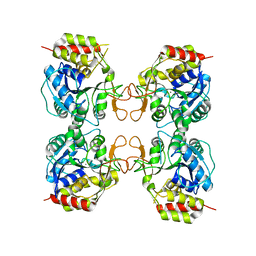 | | Crystal structure of the E. coli pyrimidine nucleoside hydrolase YeiK (Apo-form) | | Descriptor: | CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
6HT2
 
 | |
6HU5
 
 | |
2H9W
 
 | |
2O2B
 
 | |
