1D32
 
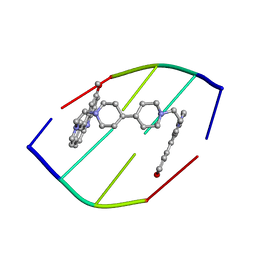 | | DRUG-INDUCED DNA REPAIR: X-RAY STRUCTURE OF A DNA-DITERCALINIUM COMPLEX | | Descriptor: | DITERCALINIUM, DNA (5'-D(*CP*GP*CP*G)-3') | | Authors: | Gao, Q, Williams, L.D, Egli, M, Rabinovich, D, Chen, S.-L, Quigley, G.J, Rich, A. | | Deposit date: | 1991-01-23 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Drug-induced DNA repair: X-ray structure of a DNA-ditercalinium complex.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
8Z9S
 
 | |
8CE9
 
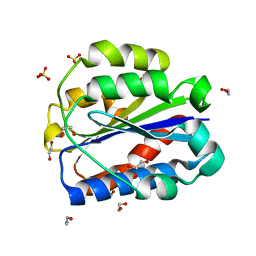 | | Crystal structure of human Cd11b I domain in C121 space group | | Descriptor: | 1,2-ETHANEDIOL, Integrin alpha-M, SULFATE ION | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of human Cd11b I domain in C121 space group
To Be Published
|
|
8CE6
 
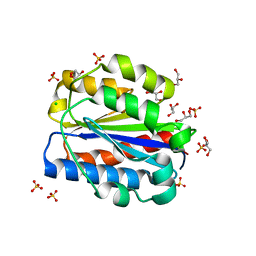 | | Crystal structure of human Cd11b I domain in P212121 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Integrin alpha-M, ... | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of human Cd11b I domain in P212121 space group
To Be Published
|
|
1D48
 
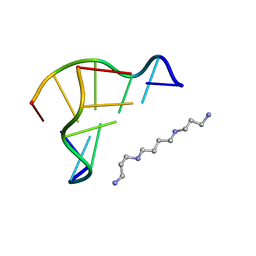 | | STRUCTURE OF THE PURE-SPERMINE FORM OF Z-DNA (MAGNESIUM FREE) AT 1 ANGSTROM RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Egli, M, Williams, L.D, Gao, Q, Rich, A. | | Deposit date: | 1991-09-11 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-A resolution.
Biochemistry, 30, 1991
|
|
154D
 
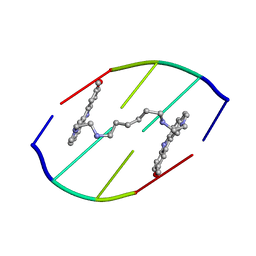 | | DNA DISTORTION IN BIS-INTERCALATED COMPLEXES | | Descriptor: | BIS-(N-ETHYLPYRIDINIUM-(3-METHOXYCARBAZOLE))HEXANE-1,6-DIAMINE, DNA (5'-D(*(CBR)P*GP*CP*G)-3') | | Authors: | Peek, M.E, Lipscomb, L.A, Bertrand, J.A, Gao, Q, Roques, B.P, Garbay-Jaureguiberry, C, Williams, L.D. | | Deposit date: | 1993-12-14 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA distortion in bis-intercalated complexes.
Biochemistry, 33, 1994
|
|
5C4W
 
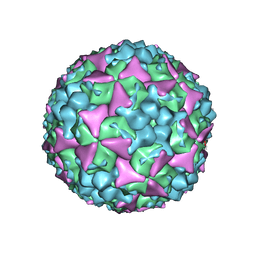 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
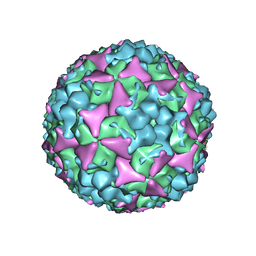 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C8C
 
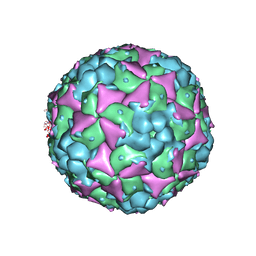 | | Crystal structure of recombinant coxsackievirus A16 capsid | | Descriptor: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-25 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
4QPG
 
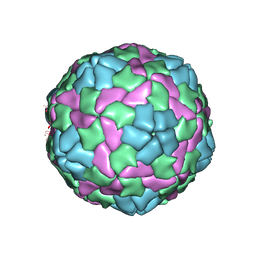 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4JGZ
 
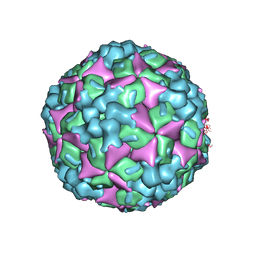 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JGY
 
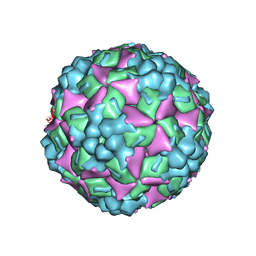 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group P4232) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, Mcauley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
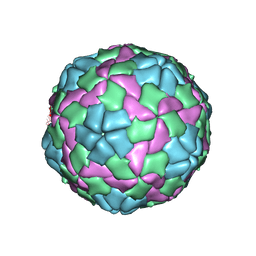 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YWO
 
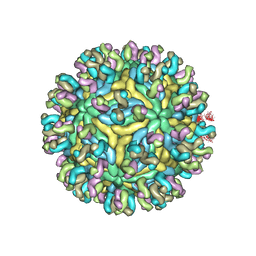 | | Structure of JEV-2F2 Fab complex | | Descriptor: | 2F2 heavy chain, 2F2 light chain, JEV E protein, ... | | Authors: | Qiu, X, Lei, Y.F, Yang, P, Gao, Q, Wang, N, Cao, L, Yuan, S, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
6AKS
 
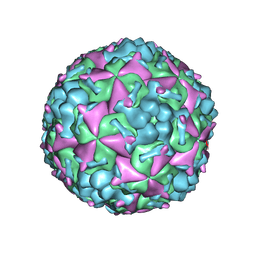 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKT
 
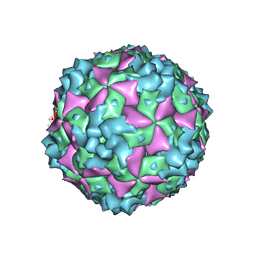 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AKU
 
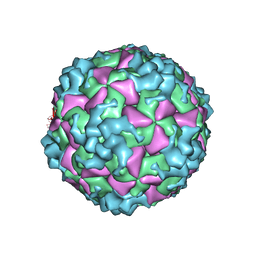 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
5YWP
 
 | | JEV-2H4 Fab complex | | Descriptor: | 2H4 heavy chain, 2H4 light chain, JEV E protein, ... | | Authors: | Qiu, X.D, Lei, Y.F, Yang, P, Gao, Q, WANG, N, Cao, L, Yuan, S, Wang, X.X, Xu, Z.K, Rao, Z.H. | | Deposit date: | 2017-11-29 | | Release date: | 2018-05-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5YWF
 
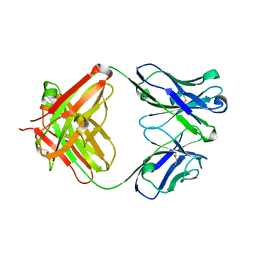 | | Crystal structure of 2H4 Fab | | Descriptor: | 2H4 heavy chain, 2H4 light chain | | Authors: | Qiu, X, Lei, Y, Yang, P, Gao, Q, Wang, N, Cao, L, Wang, X, Xu, Z.K, Rao, Z. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for neutralization of Japanese encephalitis virus by two potent therapeutic antibodies
Nat Microbiol, 3, 2018
|
|
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
