4V48
 
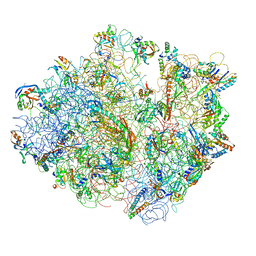 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4V47
 
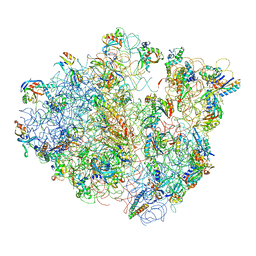 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
6RPJ
 
 | |
6XY2
 
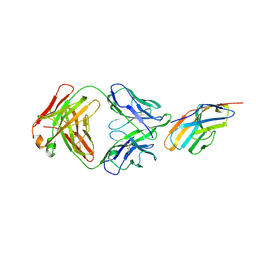 | | Crystal structure of CTLA-4 complexed with the Fab of HL32 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Heavy chain, ... | | Authors: | Gao, H, Zhou, A. | | Deposit date: | 2020-01-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of CTLA-4 complexed with a pH-sensitive cancer immunotherapeutic antibody.
Cell Discov, 6, 2020
|
|
2O0F
 
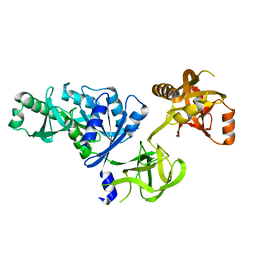 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | Descriptor: | Peptide chain release factor 3 | | Authors: | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | Deposit date: | 2006-11-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
3DG4
 
 | |
3DG2
 
 | |
3DG5
 
 | |
3DG0
 
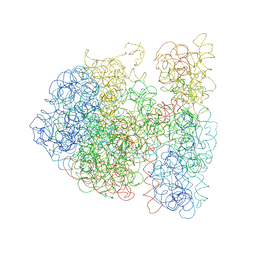 | |
6VEF
 
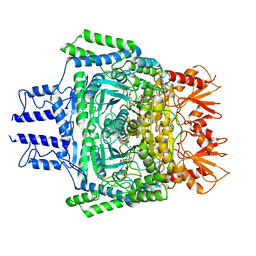 | |
7VCU
 
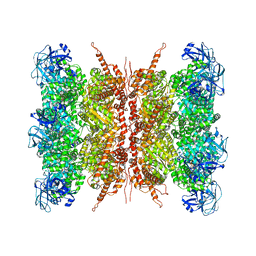 | | Human p97 double hexamer conformer I with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCT
 
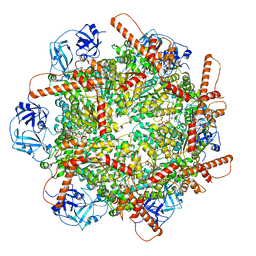 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCX
 
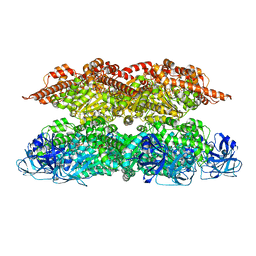 | | Human p97 single hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCS
 
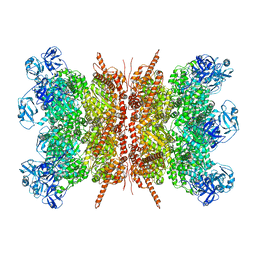 | | Human p97 double hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCV
 
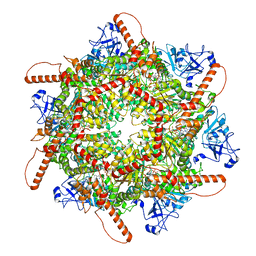 | | Human p97 single hexamer conformer I with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
6RQM
 
 | |
8AG1
 
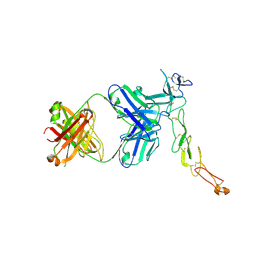 | | Crystal structure of a novel OX40 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Novel OX40 antibody heavy chain, Novel OX40 antibody light chain, ... | | Authors: | Gao, H, Zhou, A. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
4HPZ
 
 | |
7YEA
 
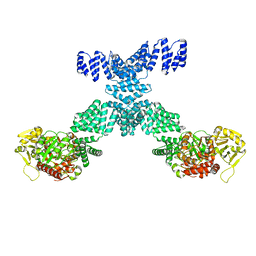 | | Human O-GlcNAc transferase Dimer | | Descriptor: | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Gao, H, Lu, P, Liu, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Cryo-EM structure of human O-GlcNAcylation enzyme pair OGT-OGA complex.
Nat Commun, 14, 2023
|
|
4A3N
 
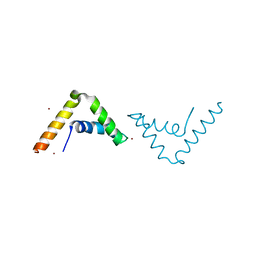 | | Crystal Structure of HMG-BOX of Human SOX17 | | Descriptor: | TRANSCRIPTION FACTOR SOX-17, ZINC ION | | Authors: | Gao, N, Gao, H, Qian, H, Si, S, Xie, Y. | | Deposit date: | 2011-10-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Human Transcription Factor Sry-Related Box 17 Binding to DNA.
Protein Pept.Lett., 20, 2013
|
|
2W36
 
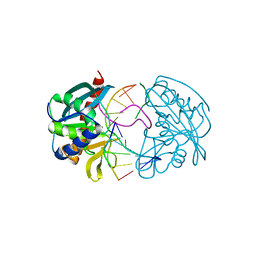 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W35
 
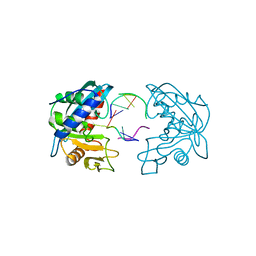 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6WG3
 
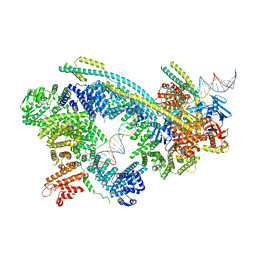 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
6WGE
 
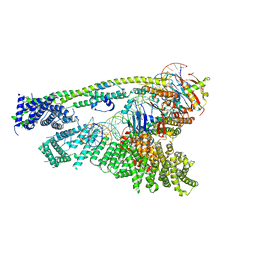 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex without STAG1 | | Descriptor: | DNA (43-MER), Double-strand-break repair protein rad21 homolog, MAGNESIUM ION, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
7YLL
 
 | | Crystal structure of TTEDbh | | Descriptor: | DNA polymerase IV, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Yan, X, Tian, L, Gao, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6000092 Å) | | Cite: | Structure and function of extreme TLS DNA polymerase TTEDbh from Thermoanaerobacter tengcongensis.
Int.J.Biol.Macromol., 253, 2023
|
|
