1BCF
 
 | | THE STRUCTURE OF A UNIQUE, TWO-FOLD SYMMETRIC, HAEM-BINDING SITE | | Descriptor: | BACTERIOFERRITIN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Frolow, F, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1993-12-06 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a unique twofold symmetric haem-binding site.
Nat.Struct.Biol., 1, 1994
|
|
3F2L
 
 | | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Frolow, F, Freeman, A, Wine, Y, Eppel, A, Shanzer, M, Stempler, S. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure analysis of the K171A mutation of N-terminal type II cohesin 1 from the cellulosomal ScaB subunit of Acetivibrio cellulolyticus
To be Published
|
|
2YEO
 
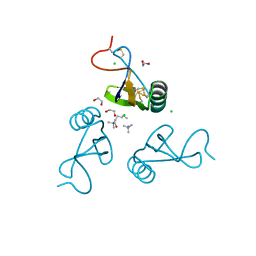 | | A39L mutation of scorpion toxin lqh-alpha-it | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-INSECT TOXIN LQHAIT, CHLORIDE ION, ... | | Authors: | Frolow, F, Kahn, R, Gurevitz, M. | | Deposit date: | 2011-03-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal Structures of Scorpion Alpha-Toxinsmutants Reveal Conformational Constraints that Dictate Preference for Mammalian Brain Voltage-Gated Na- Channels
To be Published
|
|
1DOI
 
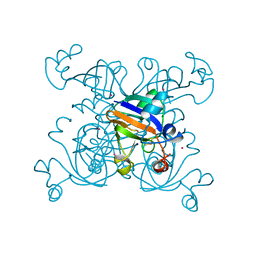 | | 2FE-2S FERREDOXIN FROM HALOARCULA MARISMORTUI | | Descriptor: | 2FE-2S FERREDOXIN, FE2/S2 (INORGANIC) CLUSTER, POTASSIUM ION | | Authors: | Frolow, F, Harel, M, Sussman, J.L, Shoham, M. | | Deposit date: | 1996-04-08 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin.
Nat.Struct.Biol., 3, 1996
|
|
4PJ1
 
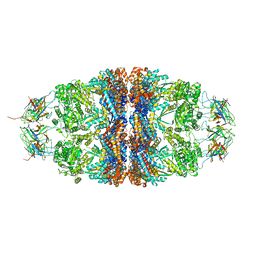 | | Crystal structure of the human mitochondrial chaperonin symmetrical 'football' complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Frolow, F, Azem, A, Nisemblat, S. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the human mitochondrial chaperonin symmetrical football complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4N2O
 
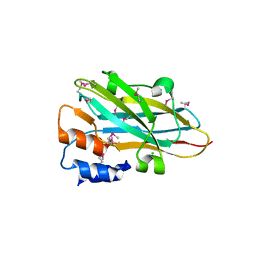 | | Structure of a novel autonomous cohesin protein from Ruminococcus flavefaciens | | Descriptor: | Autonomous cohesin, CHLORIDE ION | | Authors: | Frolow, F, Voronov-Goldman, M, Levy-Assaraf, M, Lamed, R, Bayer, E, Shimon, L. | | Deposit date: | 2013-10-05 | | Release date: | 2013-12-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | Structural characterization of a novel autonomous cohesin from Ruminococcus flavefaciens.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
2OUI
 
 | | D275P mutant of alcohol dehydrogenase from protozoa Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Shimon, L, Burstein, Y, Goihberg, E, Peretz, M, Dym, O. | | Deposit date: | 2007-02-11 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution.
Proteins, 72, 2008
|
|
4EYZ
 
 | | Crystal structure of an uncommon cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Descriptor: | 1,2-ETHANEDIOL, Cellulosome-related protein module from Ruminococcus flavefaciens that resembles papain-like cysteine peptidases | | Authors: | Frolow, F, Voronov-Goldman, M, Bayer, E, Lamed, R. | | Deposit date: | 2012-05-02 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Crystal Structure of an Uncommon Cellulosome-Related Protein Module from Ruminococcus flavefaciens That Resembles Papain-Like Cysteine Peptidases.
Plos One, 8, 2013
|
|
2ZF9
 
 | |
1QZN
 
 | | Crystal Structure Analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus | | Descriptor: | cellulosomal scaffoldin adaptor protein B | | Authors: | Frolow, F, Noach, I, Rosenheck, S, Lamed, R, Qi, X, Shimon, L.J.W, Bayer, E.A. | | Deposit date: | 2003-09-17 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements.
J.Mol.Biol., 348, 2005
|
|
3FSR
 
 | | Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NADP-dependent alcohol dehydrogenase, ... | | Authors: | Frolow, F, Greenblatt, H, Goihberg, E, Burstein, Y. | | Deposit date: | 2009-01-11 | | Release date: | 2010-01-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms
Biochemistry, 49, 2010
|
|
3FTN
 
 | | Q165E/S254K Double Mutant Chimera of alcohol dehydrogenase by exchange of the cofactor binding domain res 153-295 of T. brockii ADH by C. beijerinckii ADH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Goihberg, E, Shimon, L, Burstein, Y. | | Deposit date: | 2009-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Biochemical and structural properties of chimeras constructed by exchange of cofactor-binding domains in alcohol dehydrogenases from thermophilic and mesophilic microorganisms.
Biochemistry, 49, 2010
|
|
2I61
 
 | | Depressant anti-insect neurotoxin, LqhIT2 from Leiurus quinquestriatus hebraeus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CARBON DIOXIDE, ... | | Authors: | Frolow, F, Gurevitz, M, Karbat, I, Turkov, M, Gordon, D. | | Deposit date: | 2006-08-27 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray Structure and Mutagenesis of the Scorpion Depressant Toxin LqhIT2 Reveals Key Determinants Crucial for Activity and Anti-Insect Selectivity.
J.Mol.Biol., 366, 2007
|
|
2HUB
 
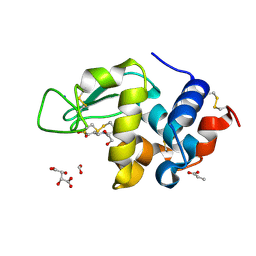 | | Structure of Hen Egg-White Lysozyme Determined from crystals grown in pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D(-)-TARTARIC ACID, ... | | Authors: | Frolow, F, Lagziel-Simis, S, Cohen-Hadar, N, Wine, Y, Freeman, A. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Monitoring influence of pH on the molecular and crystal structure of hen egg-white tetragonal lysozyme
To be Published
|
|
2HU3
 
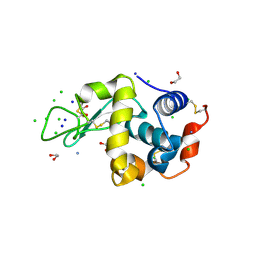 | | Parent Structure of Hen Egg White Lysozyme grown in acidic pH 4.8. Refinement for comparison with crosslinked molecules of lysozyme | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Frolow, F, Lagziel-Simis, S, Cohen-Hadar, N, Wine, Y, Freeman, A. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Monitoring influence of pH on the molecular and crystal structure of hen egg-white tetragonal lysozyme
To be Published
|
|
3L8Q
 
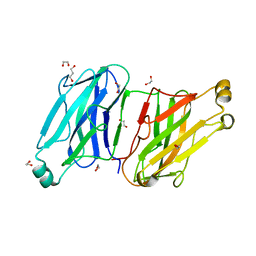 | | Structure analysis of the type II cohesin dyad from the adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2010-01-03 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Modular Arrangement of a Cellulosomal Scaffoldin Subunit Revealed from the Crystal Structure of a Cohesin Dyad
J.Mol.Biol., 399, 2010
|
|
3D05
 
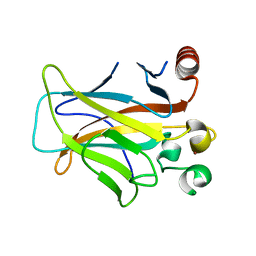 | | Human p53 core domain with hot spot mutation R249S (II) | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Suad, O, Rozenberg, H, Shimon, L.J.W, Frolow, F, Shakked, Z. | | Deposit date: | 2008-05-01 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations
J.Mol.Biol., 385, 2009
|
|
2D95
 
 | | LOW-TEMPERATURE STUDY OF THE A-DNA FRAGMENT D(GGGCGCCC) | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Eisenstein, M, Hope, H, Haran, T.E, Frolow, F, Shakked, Z, Rabinovich, D. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature study of the A-DNA fragment d(GGGCGCCC)
ACTA CRYSTALLOGR.,SECT.B, 44, 1988
|
|
7KCI
 
 | | DETERMINANTS OF REPRESSOR/OPERATOR RECOGNITION FROM THE STRUCTURE OF THE TRP OPERATOR BINDING SITE | | Descriptor: | Self-complementary deoxyoligonucleotide decamer d(CCACTAGTGG) | | Authors: | Shakked, Z, Guzikevich-Guerstein, G, Frolow, F, Rabinovich, D, Joachimiak, A, Sigler, P.B. | | Deposit date: | 1994-09-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Determinants of repressor/operator recognition from the structure of the trp operator binding site.
Nature, 368, 1994
|
|
2NVB
 
 | | Contribution of Pro275 to the Thermostability of the Alcohol Dehydrogenases (ADHs) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Goihberg, E, Tel-Or, S, Peretz, M, Frolow, F, Dym, O, Burstein, Y, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2006-11-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermal stabilization of the protozoan Entamoeba histolytica alcohol dehydrogenase by a single proline substitution
Proteins, 72, 2008
|
|
3FNK
 
 | | Crystal structure of the second type II cohesin module from the cellulosomal adaptor ScaA scaffoldin of Acetivibrio cellulolyticus | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 1,4-BUTANEDIOL, ... | | Authors: | Noach, I, Frolow, F, Bayer, E.A. | | Deposit date: | 2008-12-25 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Intermodular Linker Flexibility Revealed from Crystal Structures of Adjacent Cellulosomal Cohesins of Acetivibrio cellulolyticus
J.Mol.Biol., 391, 2009
|
|
3GHP
 
 | |
4C8X
 
 | |
6ZOI
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conknunitzin-C3 mutante | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | A lid blocking mechanism of a cone snail toxin revealed at the atomic level
To Be Published
|
|
2XDH
 
 | | Non-cellulosomal cohesin from the hyperthermophilic archaeon Archaeoglobus fulgidus | | Descriptor: | CHLORIDE ION, COHESIN, MAGNESIUM ION, ... | | Authors: | Voronov-Goldman, M, Lamed, R, Noach, I, Borovok, I, Kwiat, M, Rosenheck, S, Shimon, L.J.W, Bayer, E.A, Frolow, F. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Non-Cellulosomal Cohesin from the Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Proteins, 79, 2011
|
|
