2UWQ
 
 | |
2BIO
 
 | |
2BIP
 
 | |
2BIN
 
 | |
2BIM
 
 | |
2BIQ
 
 | |
1FY9
 
 | |
1FYA
 
 | |
2FEJ
 
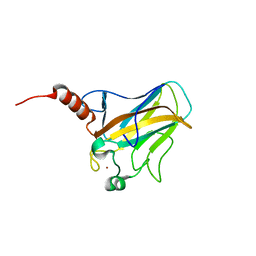 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2YBG
 
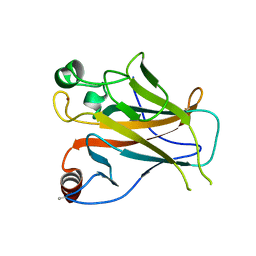 | | Structure of Lys120-acetylated p53 core domain | | Descriptor: | CELLULAR TUMOR ANTIGEN P53, ZINC ION | | Authors: | Arbely, E, Allen, M.D, Joerger, A.C, Fersht, A.R. | | Deposit date: | 2011-03-08 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acetylation of Lysine 120 of P53 Endows DNA- Binding Specificity at Effective Physiological Salt Concentration.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5LAP
 
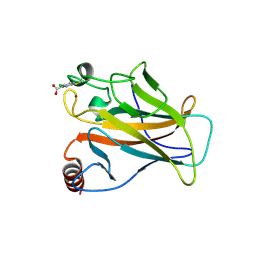 | | p53 cancer mutant Y220C with Cys182 alkylation | | Descriptor: | 5-chloranylpyrimidine-4-carboxylic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Joerger, A.C, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 2-Sulfonylpyrimidines: Mild alkylating agents with anticancer activity toward p53-compromised cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5O1E
 
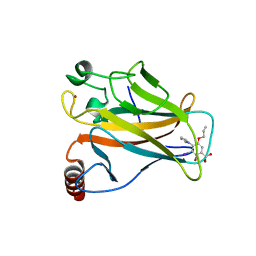 | | p53 cancer mutant Y220C im complex with compound MB577 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-prop-2-enoxy-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1A
 
 | | p53 cancer mutant Y220C in complex with compound MB240 | | Descriptor: | Cellular tumor antigen p53, GLYCEROL, ZINC ION, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1H
 
 | | p53 cancer mutant Y220C in complex with compound MB539 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-propylsulfanyl-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1G
 
 | | p53 cancer mutant Y220C in complex with compound MB487 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-(2-phenylethoxy)-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1B
 
 | | p53 cancer mutant Y220C in complex with compound MB84 | | Descriptor: | 6-(hydroxymethyl)-2,4-bis(iodanyl)-3-pyrrol-1-yl-phenol, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
1HL2
 
 | |
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1P7J
 
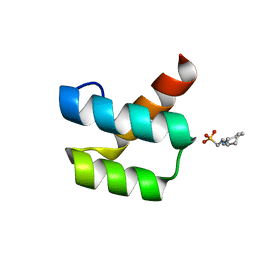 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1UOL
 
 | |
5NP0
 
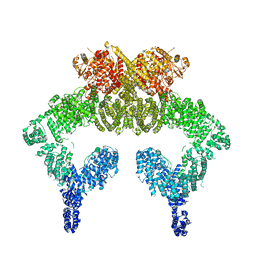 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5NP1
 
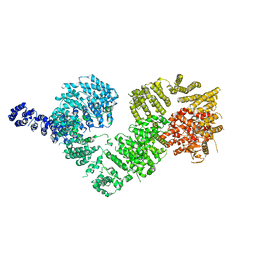 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
5O1D
 
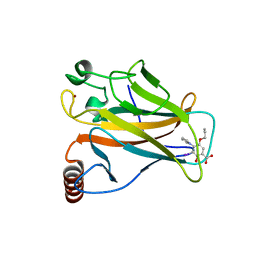 | | p53 cancer mutant Y220C in complex with compound MB481 | | Descriptor: | 3-iodanyl-2-oxidanyl-5-propoxy-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Bauer, M.R, Baud, M.G.J, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
5O1C
 
 | | p53 cancer mutant Y220C in complex with compound MB184 | | Descriptor: | 5-(4-fluorophenyl)-3-iodanyl-2-oxidanyl-4-pyrrol-1-yl-benzoic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Joerger, A.C, Baud, M.G.J, Bauer, M.R, Fersht, A.R. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Aminobenzothiazole derivatives stabilize the thermolabile p53 cancer mutant Y220C and show anticancer activity in p53-Y220C cell lines.
Eur J Med Chem, 152, 2018
|
|
