6UZC
 
 | |
5E5A
 
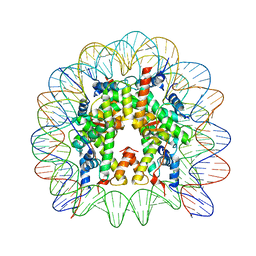 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
6NCL
 
 | |
7VS5
 
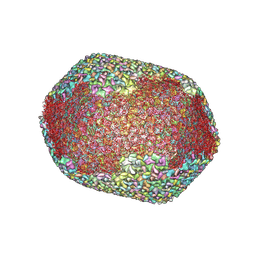 | | The expanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein, Small outer capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-25 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VRT
 
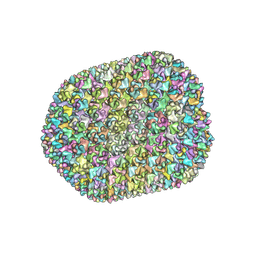 | | The unexpanded head structure of phage T4 | | Descriptor: | Capsid vertex protein, Major capsid protein | | Authors: | Fang, Q, Tang, W, Fokine, A, Mahalingam, M, Shao, Q, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6NQD
 
 | |
9KMH
 
 | | The Composite Cryo-EM Structure of the Portal Vertex of Bacteriophage FCWL1 | | Descriptor: | Adaptor protein, Connector protein, Decoration protein, ... | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-11-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
9KMG
 
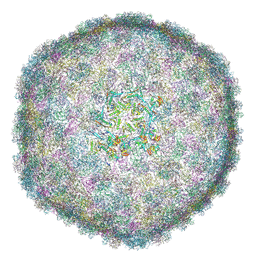 | | Cryo-EM Structure of Bacteriophage FCWL1 Capsid | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-11-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
9LBN
 
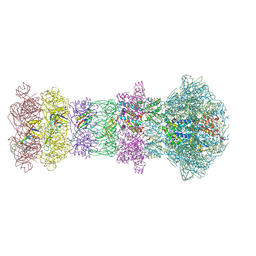 | | The composite cryo-EM structure of the head-to-tail connector and head-proximal tail components of bacteriophage phiXacJX1 | | Descriptor: | adaptor protein gp5, portal protein gp1, stopper protein gp6, ... | | Authors: | Guo, M, Wang, A, Zheng, Y, Liu, C, Shao, Q, Fang, Q. | | Deposit date: | 2025-01-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of a Xanthomonas phage: Insights into viral architecture and implications for the model phage HK97.
Structure, 33, 2025
|
|
9LW8
 
 | | Bottom cap of bacteriophage Mycofy1 mature head (C5 symmetry) | | Descriptor: | Phage capsid-like C-terminal domain-containing protein | | Authors: | Li, X, Shao, Q, Li, L, Xie, L, Ruan, Z, Fang, Q. | | Deposit date: | 2025-02-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM Reveals Structural Diversity in Prolate-headed Mycobacteriophage Mycofy1.
J.Mol.Biol., 437, 2025
|
|
9LBM
 
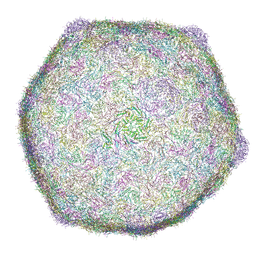 | | Cryo-EM structure of bacteriophage phiXacJX1 capsid | | Descriptor: | major capsid protein gp3 | | Authors: | Guo, M, Wang, A, Zheng, Y, Liu, C, Shao, Q, Fang, Q. | | Deposit date: | 2025-01-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of a Xanthomonas phage: Insights into viral architecture and implications for the model phage HK97.
Structure, 33, 2025
|
|
9LWA
 
 | | Bacteriophage Mycofy1 distal head-to-tail interface (C6 symmetry) | | Descriptor: | Head-to-tail stopper, Major tail protein, Terminator protein gp11 | | Authors: | Li, X, Shao, Q, Li, L, Xie, L, Ruan, Z, Fang, Q. | | Deposit date: | 2025-02-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Cryo-EM Reveals Structural Diversity in Prolate-headed Mycobacteriophage Mycofy1.
J.Mol.Biol., 437, 2025
|
|
9LW7
 
 | | Midsection of bacteriophage Mycofy1 mature head (C5 symmetry) | | Descriptor: | Phage capsid-like C-terminal domain-containing protein | | Authors: | Li, X, Shao, Q, Li, L, Xie, L, Ruan, Z, Fang, Q. | | Deposit date: | 2025-02-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM Reveals Structural Diversity in Prolate-headed Mycobacteriophage Mycofy1.
J.Mol.Biol., 437, 2025
|
|
9LW9
 
 | | Bacteriophage Mycofy1 proximal head-to-tail interface (C6 symmetry) | | Descriptor: | Adaptor protein gp8, Portal protein | | Authors: | Li, X, Shao, Q, Li, L, Xie, L, Ruan, Z, Fang, Q. | | Deposit date: | 2025-02-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Reveals Structural Diversity in Prolate-headed Mycobacteriophage Mycofy1.
J.Mol.Biol., 437, 2025
|
|
9LW6
 
 | | Top cap of bacteriophage Mycofy1 mature head (C5 symmetry) | | Descriptor: | Phage capsid-like C-terminal domain-containing protein | | Authors: | Li, X, Shao, Q, Li, L, Xie, L, Ruan, Z, Fang, Q. | | Deposit date: | 2025-02-13 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Cryo-EM Reveals Structural Diversity in Prolate-headed Mycobacteriophage Mycofy1.
J.Mol.Biol., 437, 2025
|
|
7SGS
 
 | | Cryo-EM structure of full-length MAP7 bound to the microtubule | | Descriptor: | Ensconsin, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ferro, L.S, Fang, Q, Eshun-Wilson, L, Fernandes, J, Jack, A, Farrell, D.P, Golcuk, M, Huijben, T, Costa, K, Gur, M, DiMaio, F, Nogales, E, Yildiz, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional insight into regulation of kinesin-1 by microtubule-associated protein MAP7.
Science, 375, 2022
|
|
9JJH
 
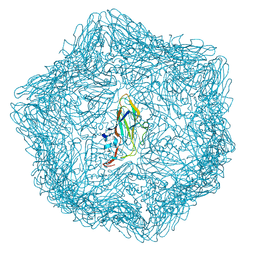 | | Cryo-EM structure of a T=1 VLP of RHDV GI.2 with N-terminal 1-37 residues truncated | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 98, 2024
|
|
9JJG
 
 | | Cryo-EM structure of RHDV GI.2 virion | | Descriptor: | Genome polyprotein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 98, 2024
|
|
9JJI
 
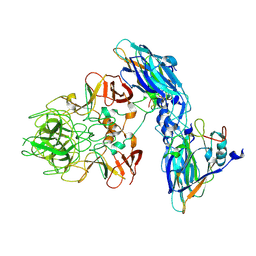 | | Local refinement of RHDV GI.2 T=1 VLP | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 98, 2024
|
|
9JJJ
 
 | | Cryo-EM structure of a T=3 VLP of RHDV GI.2 | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP.
J.Virol., 98, 2024
|
|
9JLF
 
 | | Cryo-EM Structure of Bacteriophage FCWL1 head-to-tail interface | | Descriptor: | Adaptor protein, Connector protein, Portal protein, ... | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
3EP7
 
 | | Human AdoMetDC E256Q mutant complexed with S-Adenosylmethionine methyl ester and no putrescine bound | | Descriptor: | PYRUVIC ACID, S-ADENOSYLMETHIONINE METHYL ESTER, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
3EPB
 
 | | Human AdoMetDC E256Q mutant complexed with putrescine | | Descriptor: | 1,4-DIAMINOBUTANE, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
3EP4
 
 | | Human AdoMetDC E256Q mutant with no putrescine bound | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PYRUVIC ACID, S-adenosylmethionine decarboxylase alpha chain, ... | | Authors: | Bale, S, Lopez, M.M, Makhatadze, G.I, Fang, Q, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis for Putrescine Activation of Human S-Adenosylmethionine Decarboxylase.
Biochemistry, 47, 2008
|
|
