1KLM
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1C0U
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | Descriptor: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTQ
 
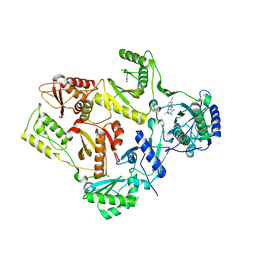 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
5ACA
 
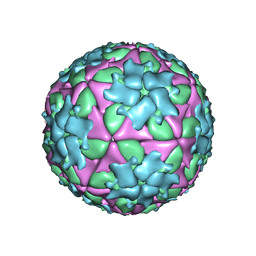 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
5AC9
 
 | | Structure-based energetics of protein interfaces guide Foot-and-Mouth disease virus vaccine design | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, PerezMartin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-14 | | Release date: | 2015-09-23 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-Based Energetics of Protein Interfaces Guide Foot-and-Mouth Disease Vaccine Design
Nat.Struct.Mol.Biol., 22, 2015
|
|
2VD8
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VD9
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2UVD
 
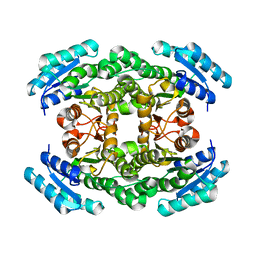 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2W8X
 
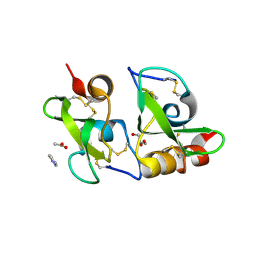 | | Structure of the tick ion-channel modulator Ra-KLP | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Paesen, G.C, Siebold, C, Dallas, M, Peers, C, Harlos, K, Nuttall, P.A, Nunn, M.A, Stuart, D.I, Esnouf, R.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-05 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Channel Modulator from the Saliva of the Brown Ear Tick Has a Highly Modified Kunitz/Bpti Structure.
J.Mol.Biol., 389, 2009
|
|
5D8A
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093F empty capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-16 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | Descriptor: | Foot and mouth disease virus, VP1, VP2, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2JCB
 
 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2JJX
 
 | | THE CRYSTAL STRUCTURE OF UMP KINASE FROM BACILLUS ANTHRACIS (BA1797) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, URIDYLATE KINASE | | Authors: | Meier, C, Carter, L.G, Mancini, E.J, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The Crystal Structure of Ump Kinase from Bacillus Anthracis (Ba1797) Reveals an Allosteric Nucleotide-Binding Site.
J.Mol.Biol., 381, 2008
|
|
2BNU
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | T-CELL RECEPTOR ALPHA CHAIN C REGION, T-CELL RECEPTOR BETA CHAIN C REGION | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, Van Der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-04-04 | | Release date: | 2005-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines.
J.Exp.Med., 201, 2005
|
|
2BNR
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, SYNTHETIC PEPTIDE, ... | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, van der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
J.Exp.Med., 201, 2005
|
|
2BNQ
 
 | | Structural and kinetic basis for heightened immunogenicity of T cell vaccines | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, SYNTHETIC PEPTIDE, ... | | Authors: | Chen, J.-L, Stewart-Jones, G, Bossi, G, Lissin, N.M, Wooldridge, L, Choi, E.M.L, Held, G, Dunbar, P.R, Esnouf, R.M, Sami, M, Boultier, J.M, Rizkallah, P.J, Renner, C, Sewell, A, van der Merwe, P.A, Jackobsen, B.K, Griffiths, G, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2005-03-31 | | Release date: | 2005-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Kinetic Basis for Heightened Immunogenicity of T Cell Vaccines
J.Exp.Med., 201, 2005
|
|
1EC4
 
 | | SOLUTION STRUCTURE OF A HEXITOL NUCLEIC ACID DUPLEX WITH FOUR CONSECUTIVE T:T BASE PAIRS | | Descriptor: | HEXITOL DODECANUCLEOTIDE | | Authors: | Lescrinier, E, Esnouf, R.M, Schraml, J, Busson, R, Herdewijn, P. | | Deposit date: | 2000-01-25 | | Release date: | 2003-04-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid
Chem.Biol., 7, 2000
|
|
1GP0
 
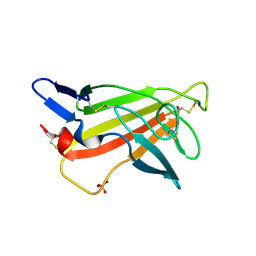 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, SULFATE ION | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
1YJD
 
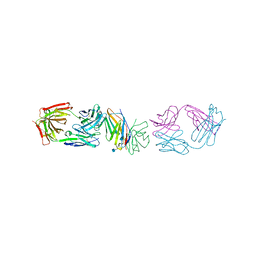 | | Crystal structure of human CD28 in complex with the Fab fragment of a mitogenic antibody (5.11A1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab fragment of 5.11A1 antibody heavy chain, Fab fragment of 5.11A1 antibody light chain, ... | | Authors: | Evans, E.J, Esnouf, R.M, Manso-Sancho, R, Gilbert, R.J.C, James, J.R, Sorensen, P, Stuart, D.I, Davis, S.J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a soluble CD28-Fab complex
Nat.Immunol., 6, 2005
|
|
1GP3
 
 | | Human IGF2R domain 11 | | Descriptor: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Esnouf, R.M, Jones, M.A, Linnell, J, Harlos, K, Hassan, A.B, Jones, E.Y. | | Deposit date: | 2001-10-29 | | Release date: | 2002-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a Functional Igf2R Fragment Determined from the Anomalous Scattering of Sulfur
Embo J., 21, 2002
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1C1B
 
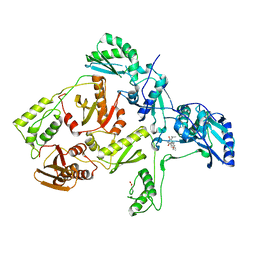 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | Descriptor: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C1C
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | Descriptor: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
