2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N08
 
 | | NMR structure of a short hydrophobic 11mer peptide in 25 mM SDS solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0I
 
 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
7S15
 
 | | GLP-1 receptor bound with Pfizer small molecule agonist | | Descriptor: | 2-[(4-{6-[(2,4-difluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-[(1-ethyl-1H-imidazol-5-yl)methyl]-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor | | Authors: | Liu, Y, Dias, J.M, Han, S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Small-Molecule Oral Agonist of the Human Glucagon-like Peptide-1 Receptor.
J.Med.Chem., 65, 2022
|
|
5KQ5
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-07-05 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated Protein Kinase (AMPK), for the Potential Treatment of Diabetic Nephropathy.
J.Med.Chem., 59, 2016
|
|
5T5T
 
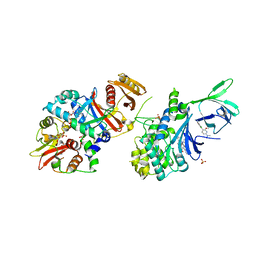 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Selective Activation of AMPK beta 1-Containing Isoforms Improves Kidney Function in a Rat Model of Diabetic Nephropathy.
J. Pharmacol. Exp. Ther., 361, 2017
|
|
8G93
 
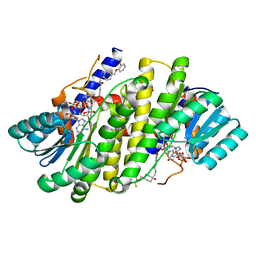 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G89
 
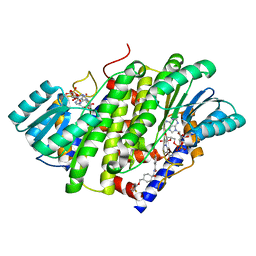 | | HSD17B13 in complex with cofactor and inhibitor | | Descriptor: | 3-fluoro-N-({(1r,4r)-4-[(2-fluorophenoxy)methyl]-1-hydroxycyclohexyl}methyl)-4-hydroxybenzamide, Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G84
 
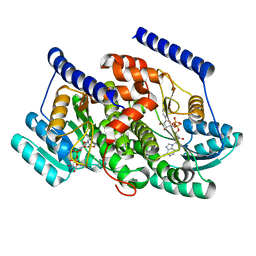 | | Crystal structures of HSD17B13 complexes | | Descriptor: | Hydroxysteroid 17-beta dehydrogenase 13, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, S. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.467 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
8G9V
 
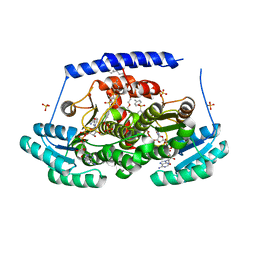 | | Crystal structures of 17-beta-hydroxysteroid dehydrogenase 13 | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 13, 4-{[2,5-dimethyl-3-(4-methylbenzene-1-sulfonyl)benzene-1-sulfonyl]amino}benzoic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Liu, S. | | Deposit date: | 2023-02-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural basis of lipid-droplet localization of 17-beta-hydroxysteroid dehydrogenase 13.
Nat Commun, 14, 2023
|
|
2NAV
 
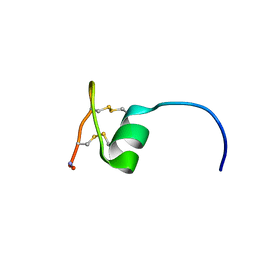 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2016-08-17 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
2NAW
 
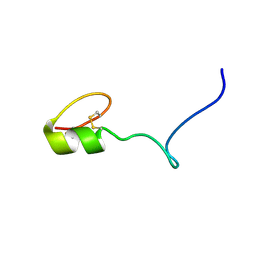 | |
6E4T
 
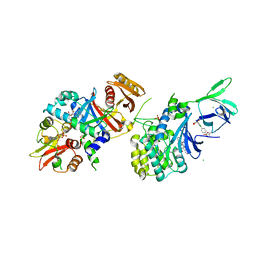 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
6E4U
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-{6-chloro-5-[6-(dimethylamino)-2-methoxypyridin-3-yl]-1H-indole-3-carbonyl}-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
6E4W
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1-O-(4,6-difluoro-5-{4-[(2S)-oxan-2-yl]phenyl}-1H-indole-3-carbonyl)-beta-D-glucopyranuronic acid, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-08 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Acyl Glucuronide Metabolites of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1 H-indole-3-carboxylic Acid (PF-06409577) and Related Indole-3-carboxylic Acid Derivatives are Direct Activators of Adenosine Monophosphate-Activated Protein Kinase (AMPK).
J. Med. Chem., 61, 2018
|
|
