1GAD
 
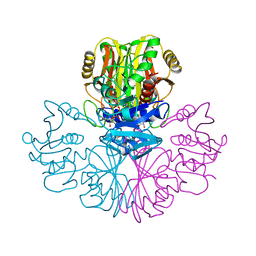 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
1GAE
 
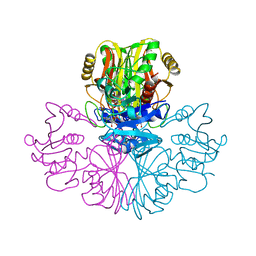 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
2BMI
 
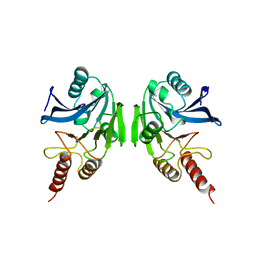 | | METALLO-BETA-LACTAMASE | | Descriptor: | PROTEIN (CLASS B BETA-LACTAMASE), SODIUM ION, ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-17 | | Release date: | 1998-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the ZnII beta-lactamase from Bacteroides fragilis in an orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1BVT
 
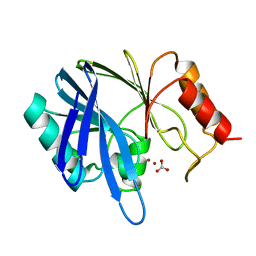 | | METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS 569/H/9 | | Descriptor: | BICARBONATE ION, PROTEIN (BETA-LACTAMASE), ZINC ION | | Authors: | Carfi, A, Duee, E, Dideberg, O. | | Deposit date: | 1998-09-18 | | Release date: | 1998-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A resolution structure of the zinc (II) beta-lactamase from Bacillus cereus.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1DXK
 
 | |
1BMC
 
 | | STRUCTURE OF A ZINC METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Carfi, A, Pares, S, Duee, E, Dideberg, O. | | Deposit date: | 1995-06-16 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold.
EMBO J., 14, 1995
|
|
1QME
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2X, SULFATE ION | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Penicillin-Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance.
J.Mol.Biol., 299, 2000
|
|
1QMF
 
 | | PENICILLIN-BINDING PROTEIN 2X (PBP-2X) ACYL-ENZYME COMPLEX | | Descriptor: | 2-[CARBOXY-(2-FURAN-2-YL-2-METHOXYIMINO-ACETYLAMINO)-METHYL]-5-METHYL-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, CEFUROXIME (OCT-3-ENE FORM), PENICILLIN-BINDING PROTEIN 2X | | Authors: | Gordon, E.J, Mouz, N, Duee, E, Dideberg, O. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Penicillin Binding Protein 2X from Streptococcus Pneumoniae and its Acyl-Enzyme Form: Implication in Drug Resistance
J.Mol.Biol., 299, 2000
|
|
1DXH
 
 | | Catabolic ornithine carbamoyltransferase from Pseudomonas aeruginosa | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, SULFATE ION | | Authors: | Sainz, G, Vicat, J, Kahn, R, Duee, E, Tricot, C, Stalon, V, Dideberg, O. | | Deposit date: | 2000-01-05 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Allosteric Active Form of Catabolic Ornithine Carbamoyltransferase from Pseudomonas Aeruginosa
To be Published
|
|
1FDN
 
 | | REFINED CRYSTAL STRUCTURE OF THE 2[4FE-4S] FERREDOXIN FROM CLOSTRIDIUM ACIDURICI AT 1.84 ANGSTROMS RESOLUTION | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Duee, E, Fanchon, E, Vicat, J, Sieker, L.C, Meyer, J, Moulis, J-M. | | Deposit date: | 1994-03-31 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Refined crystal structure of the 2[4Fe-4S] ferredoxin from Clostridium acidurici at 1.84 A resolution.
J.Mol.Biol., 243, 1994
|
|
1H6N
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Sainz, G, Jaquinod, M, Gagnon, J, Jouve, H.M. | | Deposit date: | 2001-06-20 | | Release date: | 2003-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High Resolution Structure and Biochemical Properties of a Recombinant Proteus Mirabilis Catalase Depleted in Iron.
Proteins: Struct.,Funct., Genet., 50, 2003
|
|
1H7K
 
 | | Formation of a tyrosyl radical intermediate in Proteus mirabilis catalase by directed mutagenesis and consequences for nucleotide reactivity | | Descriptor: | ACETATE ION, CATALASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Andreoletti, P, Gambarelli, S, Gaillard, J, Sainz, G, Stojanoff, V, Jouve, H.M. | | Deposit date: | 2001-07-08 | | Release date: | 2004-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formation of a Tyrosyl Radical Intermediate in Proteus Mirabilis Catalase by Directed Mutagenesis and Consequences for Nucleotide Reactivity.
Biochemistry, 40, 2001
|
|
3GPD
 
 | |
5DPX
 
 | | 1,2,4-Triazole-3-thione compounds as inhibitors of L1, di-zinc metallo-beta-lactamases. | | Descriptor: | 5-(2-methylphenyl)-3H-1,2,4-triazole-3-thione, Metallo-beta-lactamase L1 type 3, SULFATE ION, ... | | Authors: | Nauton, L, Garau, G, Khan, R, Dideberg, O. | | Deposit date: | 2015-09-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases.
ChemMedChem, 12, 2017
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
1PPI
 
 | | THE ACTIVE CENTER OF A MAMMALIAN ALPHA-AMYLASE. THE STRUCTURE OF THE COMPLEX OF A PANCREATIC ALPHA-AMYLASE WITH A CARBOHYDRATE INHIBITOR REFINED TO 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose, ALPHA-AMYLASE, CALCIUM ION, ... | | Authors: | Qian, M, Haser, R, Payan, F. | | Deposit date: | 1994-02-22 | | Release date: | 1995-05-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active center of a mammalian alpha-amylase. Structure of the complex of a pancreatic alpha-amylase with a carbohydrate inhibitor refined to 2.2-A resolution.
Biochemistry, 33, 1994
|
|
1X8H
 
 | |
1DYP
 
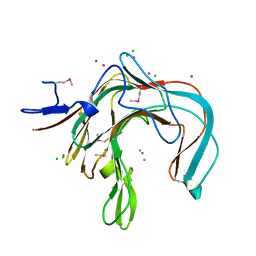 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
1PYY
 
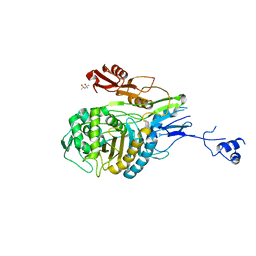 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
2CAH
 
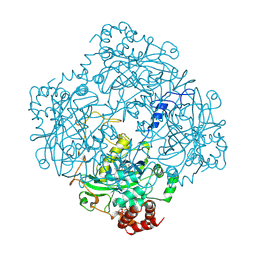 | |
1BVN
 
 | | PIG PANCREATIC ALPHA-AMYLASE IN COMPLEX WITH THE PROTEINACEOUS INHIBITOR TENDAMISTAT | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE), ... | | Authors: | Machius, M, Wiegand, G, Epp, O, Huber, R. | | Deposit date: | 1998-09-16 | | Release date: | 1998-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of porcine pancreatic alpha-amylase in complex with the microbial inhibitor Tendamistat.
J.Mol.Biol., 247, 1995
|
|
1JFH
 
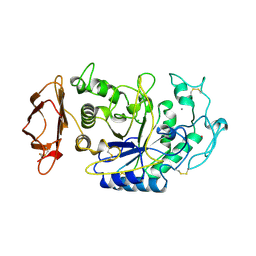 | |
1HX0
 
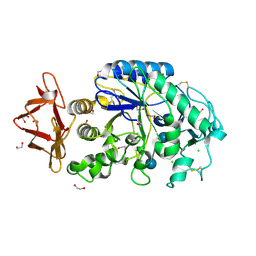 | | Structure of pig pancreatic alpha-amylase complexed with the "truncate" acarbose molecule (pseudotrisaccharide) | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ALPHA AMYLASE (PPA), ... | | Authors: | Qian, M, Payan, F. | | Deposit date: | 2001-01-11 | | Release date: | 2001-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Enzyme-catalyzed condensation reaction in a mammalian alpha-amylase. High-resolution structural analysis of an enzyme-inhibitor complex
Biochemistry, 40, 2001
|
|
1X8I
 
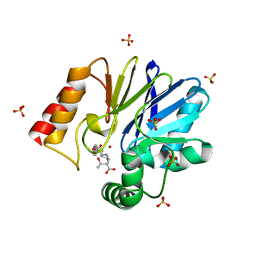 | | Crystal Structure of the Zinc Carbapenemase CphA in Complex with the Antibiotic Biapenem | | Descriptor: | 5H-PYRAZOLO(1,2-A)(1,2,4)TRIAZOL-4-IUM, 6-((2-CARBOXY-6-(1-HYDROXYETHYL)-4-METHYL-7-OXO-1-AZABICYCLO(3.2.0)HEPT-2-EN-3-YL)THIO)-6,7-DIHYDRO-, HYDROXIDE, ... | | Authors: | Garau, G, Dideberg, O. | | Deposit date: | 2004-08-18 | | Release date: | 2004-12-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Metallo-beta-lactamase Enzyme in Action: Crystal Structures of the Monozinc Carbapenemase CphA and its Complex with Biapenem
J.Mol.Biol., 345, 2005
|
|
1X8G
 
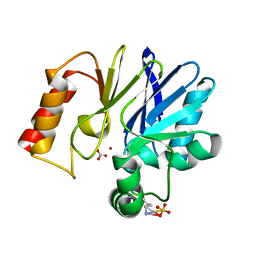 | |
