1C05
 
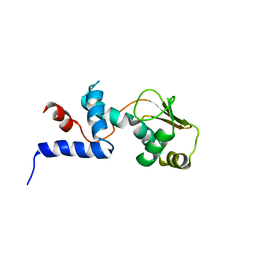 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RIBOSOMAL PROTEIN S4 DELTA 41 | | Authors: | Markus, M.A, Gerstner, R.B, Draper, D.E, Torchia, D.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases.
J.Mol.Biol., 292, 1999
|
|
1C06
 
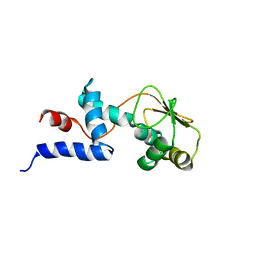 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (ENSEMBLE OF 16 STRUCTURES) | | Descriptor: | RIBOSOMAL PROTEIN S4 DELTA 41 | | Authors: | Markus, M.A, Gerstner, R.B, Draper, D.E, Torchia, D.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases.
J.Mol.Biol., 292, 1999
|
|
1ACI
 
 | |
2FOW
 
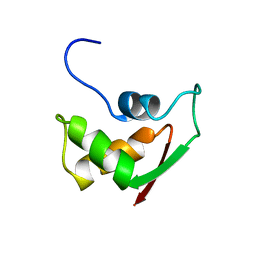 | | THE RNA BINDING DOMAIN OF RIBOSOMAL PROTEIN L11: THREE-DIMENSIONAL STRUCTURE OF THE RNA-BOUND FORM OF THE PROTEIN, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL PROTEIN L11 | | Authors: | Hinck, A.P, Markus, M.A, Huang, S, Grzesiek, S, Kustanovich, I, Draper, D.E, Torchia, D.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA.
J.Mol.Biol., 274, 1997
|
|
1FOX
 
 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1FOW
 
 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1Y39
 
 | | Co-evolution of protein and RNA structures within a highly conserved ribosomal domain | | Descriptor: | 50S ribosomal protein L11, 58 Nucleotide Ribosomal 23S RNA Domain, COBALT (III) ION, ... | | Authors: | Dunstan, M.S, GuhaThakurta, D, Draper, D.E, Conn, G.L. | | Deposit date: | 2004-11-24 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Coevolution of Protein and RNA Structures within a Highly Conserved Ribosomal Domain
Chem.Biol., 12, 2005
|
|
2E36
 
 | | L11 with SANS refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E34
 
 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E35
 
 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
1HC8
 
 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL 23S RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
J.Mol.Biol., 318, 2002
|
|
1QA6
 
 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a conserved ribosomal protein-RNA complex.
Science, 284, 1999
|
|
1FOY
 
 | | THE RNA BINDING DOMAIN OF RIBOSOMAL PROTEIN L11: THREE-DIMENSIONAL STRUCTURE OF THE RNA-BOUND FORM OF THE PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RIBOSOMAL PROTEIN L11 | | Authors: | Hinck, A.P, Markus, M.A, Huang, S, Grzesiek, S, Kustanovich, I, Draper, D.E, Torchia, D.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA.
J.Mol.Biol., 274, 1997
|
|
2H8W
 
 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5E54
 
 | | Two apo structures of the adenine riboswitch aptamer domain determined using an X-ray free electron laser | | Descriptor: | MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
1OLN
 
 | | Model for thiostrepton antibiotic binding to L11 substrate from 50S ribosomal RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RNA, THIOSTREPTON | | Authors: | Lentzen, G, Klinck, R, Matassova, N, Aboul-Ela, F, Murchie, A.I.H. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-11 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Basis for Contrasting Activities of Ribosome Binding Thiazole Antibiotics
Chem.Biol., 10, 2003
|
|
1MMS
 
 | | Crystal structure of the ribosomal PROTEIN L11-RNA complex | | Descriptor: | 23S RIBOSOMAL RNA, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Wimberly, B.T, Guymon, R, Mccutcheon, J.P, White, S.W, Ramakrishnan, V. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A detailed view of a ribosomal active site: the structure of the L11-RNA complex.
Cell(Cambridge,Mass.), 97, 1999
|
|
1QD7
 
 | | PARTIAL MODEL FOR 30S RIBOSOMAL SUBUNIT | | Descriptor: | CENTRAL FRAGMENT OF 16 S RNA, END FRAGMENT OF 16 S RNA, S15 RIBOSOMAL PROTEIN, ... | | Authors: | Clemons Jr, W.M, May, J.L.C, Wimberly, B.T, McCutcheon, J.P, Capel, M.S, Ramakrishnan, V. | | Deposit date: | 1999-07-09 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution.
Nature, 400, 1999
|
|
1FKA
 
 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
1EG0
 
 | | FITTING OF COMPONENTS WITH KNOWN STRUCTURE INTO AN 11.5 A CRYO-EM MAP OF THE E.COLI 70S RIBOSOME | | Descriptor: | FORMYL-METHIONYL-TRNA, FRAGMENT OF 16S RRNA HELIX 23, FRAGMENT OF 23S RRNA, ... | | Authors: | Gabashvili, I.S, Agrawal, R.K, Spahn, C.M.T, Grassucci, R.A, Svergun, D.I, Frank, J, Penczek, P. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Solution structure of the E. coli 70S ribosome at 11.5 A resolution.
Cell(Cambridge,Mass.), 100, 2000
|
|
