5DCN
 
 | | Crystal structure of LC3 in complex with TECPR2 LIR | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Stadel, D, Huber, J, Dotsch, V, Rogov, V.V, Behrends, C, Akutsu, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export.
Mol.Cell, 60, 2015
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
9VG5
 
 | | NMR Solution Structure of the Monomeric Catalytic C-terminal Lobe of the E6AP HECT E3 Ubiquitin Ligase | | Descriptor: | Ubiquitin-protein ligase E3A | | Authors: | Dag, C, Lambert, M, Lohr, F, Lee, W, Kazar, A.E, Dotsch, V. | | Deposit date: | 2025-06-12 | | Release date: | 2025-06-25 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of the Monomeric Catalytic C-terminal Lobe of the E6AP HECT E3 Ubiquitin Ligase
To Be Published
|
|
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
6FGS
 
 | | Solution structure of p300Taz2-p73TA1 | | Descriptor: | Histone acetyltransferase p300,Tumor protein p73, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
8RCI
 
 | | Human p53 DNA-binding domain bound to DARPin C10 | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DARPin C10, ... | | Authors: | Balourdas, D.I, Muenick, P, Strubel, A, Knapp, S, Dotsch, V, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-12-06 | | Release date: | 2024-12-18 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DARPin-induced reactivation of p53 in HPV-positive cells.
Nat.Struct.Mol.Biol., 32, 2025
|
|
1NFA
 
 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
9GFO
 
 | | iASPP-CTD fusion to p63 peptide | | Descriptor: | Tumor protein 63,RelA-associated inhibitor | | Authors: | Chaikuad, A, Akutsu, M, Lotz, R, Osterburg, C, Knapp, S, Dotsch, V. | | Deposit date: | 2024-08-12 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Alternative splicing in the DBD linker region of p63 modulates binding to DNA and iASPP in vitro.
Cell Death Dis, 16, 2025
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
5HOB
 
 | | p73 homo-tetramerization domain mutant I | | Descriptor: | MAGNESIUM ION, Tumor protein p73 | | Authors: | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Knapp, S, Dotsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.220013 Å) | | Cite: | Structural basis of p63/p73 hetero-tetramerization
To Be Published
|
|
5HOC
 
 | | p73 homo-tetramerization domain mutant II | | Descriptor: | Tumor protein p73 | | Authors: | Coutandin, D, Krojer, T, Salah, E, Mathea, S, Sumyk, M, Knapp, S, Dotsch, V. | | Deposit date: | 2016-01-19 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.36007786 Å) | | Cite: | Mechanism of TAp73 inhibition by Delta Np63 and structural basis of p63/p73 hetero-tetramerization.
Cell Death Differ., 23, 2016
|
|
1CI5
 
 | | GLYCAN-FREE MUTANT ADHESION DOMAIN OF HUMAN CD58 (LFA-3) | | Descriptor: | PROTEIN (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3(CD58)) | | Authors: | Sun, Z.Y.J, Dotsch, V, Kim, M, Li, J, Reinherz, E.L, Wagner, G. | | Deposit date: | 1999-04-07 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies.
EMBO J., 18, 1999
|
|
1EYF
 
 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | Descriptor: | ADA REGULATORY PROTEIN, ZINC ION | | Authors: | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | Deposit date: | 2000-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
4A9Z
 
 | | CRYSTAL STRUCTURE OF HUMAN P63 TETRAMERIZATION DOMAIN | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, TUMOR PROTEIN 63 | | Authors: | Muniz, J.R.C, Coutandin, D, Salah, E, Chaikuad, A, Vollmar, M, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, von Delft, F, Knapp, S. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of Human P63 Tetramerization Domain
To be Published
|
|
6FGN
 
 | | Solution Structure of p300Taz2-p63TA | | Descriptor: | Histone acetyltransferase p300,Tumor protein 63, ZINC ION | | Authors: | Gebel, J, Kazemi, S, Lohr, F, Guntert, P, Dotsch, V. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Regulation of the Activity in the p53 Family Depends on the Organization of the Transactivation Domain.
Structure, 26, 2018
|
|
2AYY
 
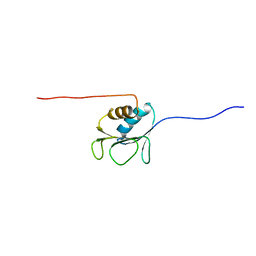 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
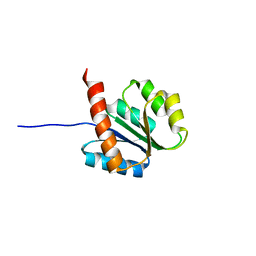 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
1ZL8
 
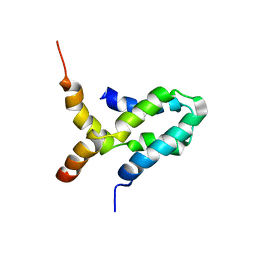 | | NMR structure of L27 heterodimer from C. elegans Lin-7 and H. sapiens Lin-2 scaffold proteins | | Descriptor: | LIN-7, Peripheral plasma membrane protein CASK | | Authors: | Petrosky, K.Y, Ou, H.D, Lohr, F, Dotsch, V, Lim, W.A. | | Deposit date: | 2005-05-05 | | Release date: | 2005-09-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A General Model for Preferential Hetero-oligomerization of LIN-2/7 Domains: Mechanism Underlying Directed Assembly of Supramolecular Signaling Complexes
J.Biol.Chem., 280, 2005
|
|
2AYX
 
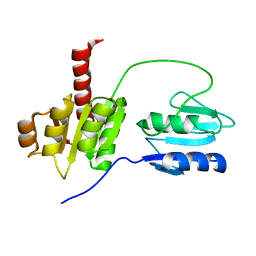 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
5N6R
 
 | | Solution structure of the Dbl-homology domain of Bcr-Abl | | Descriptor: | Breakpoint cluster region protein | | Authors: | Reckel, S, Lohr, F, Buchner, L, Guntert, P, Dotsch, V, Hantschel, O. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and functional dissection of the DH and PH domains of oncogenic Bcr-Abl tyrosine kinase.
Nat Commun, 8, 2017
|
|
6RU6
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with monophosphorylated p63 PAD1P peptide | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase I isoform delta, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RU7
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with double phosphorylated p63 PAD2P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6RU8
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with triple phosphorylated p63 PAD3P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
2K2Q
 
 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
2LIW
 
 | | NMR structure of HMG-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, 4'-PHOSPHOPANTETHEINE, CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-21 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
