4TGL
 
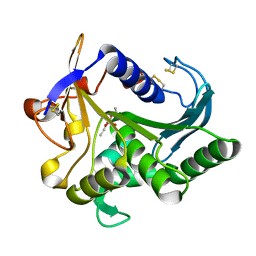 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
6INS
 
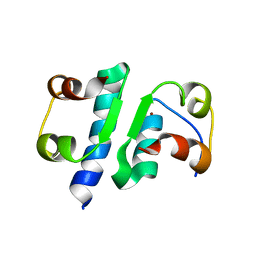 | | X-RAY ANALYSIS OF THE SINGLE CHAIN B29-A1 PEPTIDE-LINKED INSULIN MOLECULE. A COMPLETELY INACTIVE ANALOGUE | | Descriptor: | INSULIN, ZINC ION | | Authors: | Derewenda, U, Derewenda, Z, Dodson, E.J, Dodson, G.G, Bing, X, Markussen, J. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue.
J.Mol.Biol., 220, 1991
|
|
1KTJ
 
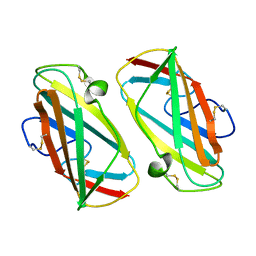 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
4GUE
 
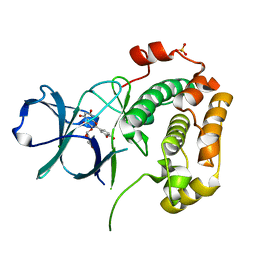 | | Structure of N-terminal kinase domain of RSK2 with flavonoid glycoside quercitrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, MAGNESIUM ION, Ribosomal protein S6 kinase alpha-3, ... | | Authors: | Derewenda, U, Utepbergenov, D, Szukalska, G, Derewenda, Z.S. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of quercitrin as an inhibitor of the p90 S6 ribosomal kinase (RSK): structure of its complex with the N-terminal domain of RSK2 at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1TIB
 
 | |
1TIC
 
 | | CONFORMATIONAL LABILITY OF LIPASES OBSERVED IN THE ABSENCE OF AN OIL-WATER INTERFACE: CRYSTALLOGRAPHIC STUDIES OF ENZYMES FROM THE FUNGI HUMICOLA LANUGINOSA AND RHIZOPUS DELEMAR | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Green, R, Joerger, R, Haas, M.J, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational lability of lipases observed in the absence of an oil-water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar.
J.Lipid Res., 35, 1994
|
|
2V71
 
 | | Coiled-coil region of NudEL | | Descriptor: | NUCLEAR DISTRIBUTION PROTEIN NUDE-LIKE 1 | | Authors: | Derewenda, U, Cooper, D.R, Kim, M.H, Derewenda, Z.S. | | Deposit date: | 2007-07-25 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Structure of the Coiled-Coil Domain of Ndel1 and the Basis of its Interaction with Lis1, the Causal Protein of Miller-Dieker Lissencephaly.
Structure, 15, 2007
|
|
1TIA
 
 | | AN UNUSUAL BURIED POLAR CLUSTER IN A FAMILY OF FUNGAL LIPASES | | Descriptor: | LIPASE | | Authors: | Derewenda, U, Swenson, L, Yamaguchi, S, Wei, Y, Derewenda, Z.S. | | Deposit date: | 1993-12-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unusual buried polar cluster in a family of fungal lipases.
Nat.Struct.Biol., 1, 1994
|
|
3FHK
 
 | | Crystal structure of APC1446, B.subtilis YphP disulfide isomerase | | Descriptor: | SULFATE ION, UPF0403 protein yphP | | Authors: | Derewenda, U, Boczek, T, Cooper, D.R, Yu, M, Hung, L, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-09 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif .
Biochemistry, 48, 2009
|
|
1R6F
 
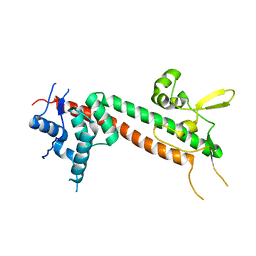 | | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague | | Descriptor: | Virulence-associated V antigen | | Authors: | Derewenda, U, Mateja, A, Devedjiev, Y, Routzahn, K.M, Evdokimov, A.G, Derewenda, Z.S, Waugh, D.S. | | Deposit date: | 2003-10-15 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague
Structure, 12, 2004
|
|
1XAS
 
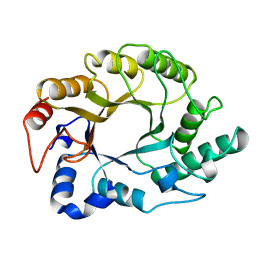 | | CRYSTAL STRUCTURE, AT 2.6 ANGSTROMS RESOLUTION, OF THE STREPTOMYCES LIVIDANS XYLANASE A, A MEMBER OF THE F FAMILY OF BETA-1,4-D-GLYCANSES | | Descriptor: | 1,4-BETA-D-XYLAN XYLANOHYDROLASE | | Authors: | Derewenda, U, Derewenda, Z.S. | | Deposit date: | 1994-05-31 | | Release date: | 1995-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure, at 2.6-A resolution, of the Streptomyces lividans xylanase A, a member of the F family of beta-1,4-D-glycanases.
J.Biol.Chem., 269, 1994
|
|
1XCG
 
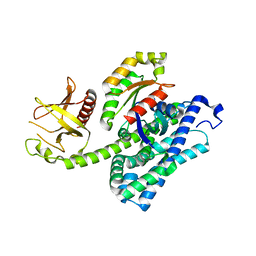 | | Crystal Structure of Human RhoA in complex with DH/PH fragment of PDZRHOGEF | | Descriptor: | Rho guanine nucleotide exchange factor 11, Transforming protein RhoA | | Authors: | Derewenda, U, Oleksy, A, Stevenson, A.S, Korczynska, J, Dauter, Z, Somlyo, A.P, Otlewski, J, Somlyo, A.V, Derewenda, Z.S. | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle
Structure, 12, 2004
|
|
5IRC
 
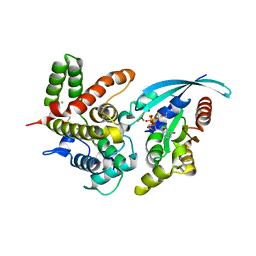 | | p190A GAP domain complex with RhoA | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Derewenda, U, Derewenda, Z. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-17 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Deciphering the Molecular and Functional Basis of RHOGAP Family Proteins: A SYSTEMATIC APPROACH TOWARD SELECTIVE INACTIVATION OF RHO FAMILY PROTEINS.
J.Biol.Chem., 291, 2016
|
|
4QAZ
 
 | |
4QB0
 
 | |
3QOR
 
 | |
5E2X
 
 | | The crystal structure of the C-terminal domain of Ebola (Tai Forest) nucleoprotein | | Descriptor: | NONAETHYLENE GLYCOL, NP | | Authors: | Baker, L.E, Handing, K.B, Derewenda, U, Utepbergenov, D, Derewenda, Z.S. | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5DSD
 
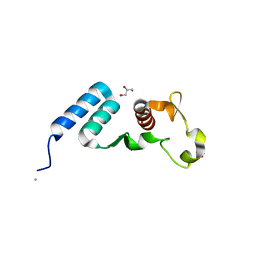 | | The crystal structure of the C-terminal domain of Ebola (Bundibugyo) nucleoprotein | | Descriptor: | CHLORIDE ION, GLYCEROL, Nucleoprotein | | Authors: | Baker, L, Handing, K.B, Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Molecular architecture of the nucleoprotein C-terminal domain from the Ebola and Marburg viruses.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6B6Z
 
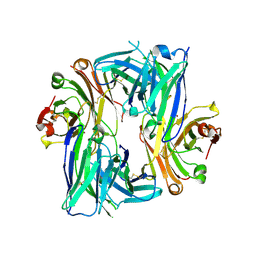 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Fab Heavy Chain, Apo Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
5TGL
 
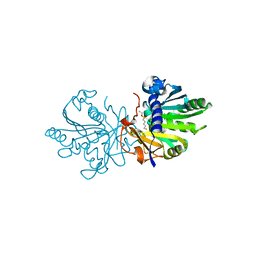 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
1BWQ
 
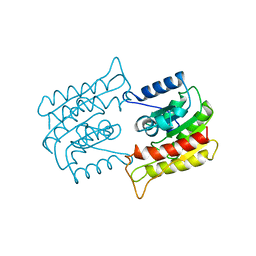 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1BWP
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1BWR
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
4EL9
 
 | | Structure of N-terminal kinase domain of RSK2 with afzelin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Ribosomal protein S6 kinase alpha-3 | | Authors: | Utepbergenov, D, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2012-04-10 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Insights into the Inhibition of the p90 Ribosomal S6 Kinase (RSK) by the Flavonol Glycoside SL0101 from the 1.5 A Crystal Structure of the N-Terminal Domain of RSK2 with Bound Inhibitor.
Biochemistry, 51, 2012
|
|
5W2B
 
 | | Crystal structure of C-terminal domain of Ebola (Reston) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
