2GCH
 
 | |
1YQV
 
 | | The crystal structure of the antibody Fab HyHEL5 complex with lysozyme at 1.7A resolution | | Descriptor: | Hen Egg White Lysozyme, HyHEL-5 Antibody Heavy Chain, HyHEL-5 Antibody Light Chain | | Authors: | Cohen, G.H, Silverton, E.W, Padlan, E.A, Dyda, F, Wibbenmeyer, J.A, Wilson, R.C, Davies, D.R. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Water molecules in the antibody-antigen interface of the structure of the Fab HyHEL-5-lysozyme complex at 1.7 A resolution: comparison with results from isothermal titration calorimetry.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2MCP
 
 | |
3SKU
 
 | | Herpes simplex virus glycoprotein D bound to the human receptor nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D, ... | | Authors: | Di Giovine, P, Settembre, E.C, Bhargava, A.K, Luftig, M.A, Lou, H, Cohen, G.H, Eisenberg, R.J, Krummenacher, C, Carfi, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of herpes simplex virus glycoprotein d bound to the human receptor nectin-1.
Plos Pathog., 7, 2011
|
|
1MCP
 
 | | PHOSPHOCHOLINE BINDING IMMUNOGLOBULIN FAB MC/PC603. AN X-RAY DIFFRACTION STUDY AT 2.7 ANGSTROMS | | Descriptor: | IGA-KAPPA MCPC603 FAB (HEAVY CHAIN), IGA-KAPPA MCPC603 FAB (LIGHT CHAIN), SULFATE ION | | Authors: | Satow, Y, Cohen, G.H, Padlan, E.A, Davies, D.R. | | Deposit date: | 1984-07-09 | | Release date: | 1985-01-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A.
J.Mol.Biol., 190, 1986
|
|
3NWA
 
 | | Glycoprotein B from Herpes simplex virus type 1, W174R mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2629 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NWF
 
 | | Glycoprotein B from Herpes simplex virus type 1, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B, MESO-ERYTHRITOL, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NW8
 
 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, high-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75915 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
3NWD
 
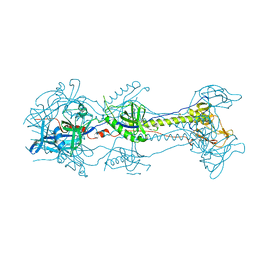 | | Glycoprotein B from Herpes simplex virus type 1, Y179S mutant, low-pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Stampfer, S.D, Lou, H, Cohen, G.H, Eisenberg, R.J, Heldwein, E.E. | | Deposit date: | 2010-07-09 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8803 Å) | | Cite: | Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1.
J.Virol., 84, 2010
|
|
4BOM
 
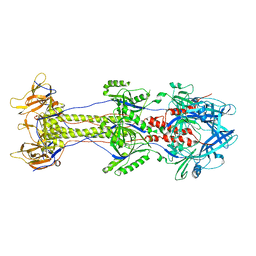 | | Structure of herpesvirus fusion glycoprotein B-bilayer complex revealing the protein-membrane and lateral protein-protein interaction | | Descriptor: | ENVELOPE GLYCOPROTEIN B | | Authors: | Maurer, U.E, Zeev-Ben-Mordehai, Z, Pandurangan, A.P, Cairns, T.M, Hannah, B.P, Whitbeck, J.C, Eisenberg, R.J, Cohen, G.H, Topf, M, Huiskonen, J.T, Grunewald, K. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (27 Å) | | Cite: | The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction.
Structure, 21, 2013
|
|
2C36
 
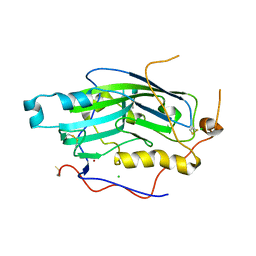 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | Descriptor: | CHLORIDE ION, GLYCOPROTEIN D HSV-1, ZINC ION, ... | | Authors: | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry.
EMBO J., 24, 2005
|
|
2C3A
 
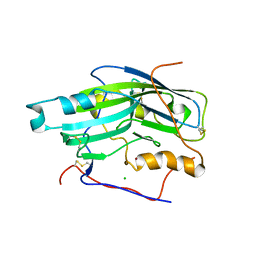 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCOPROTEIN D, ... | | Authors: | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Unliganded Hsv Gd Reveals a Mechanism for Receptor-Mediated Activation of Virus Entry.
Embo J., 24, 2005
|
|
1L2G
 
 | | Structure of a C-terminally truncated form of glycoprotein D from HSV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacher, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2002-02-21 | | Release date: | 2003-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
1JMA
 
 | | CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS GLYCOPROTEIN D BOUND TO THE CELLULAR RECEPTOR HVEA/HVEM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN D, HERPESVIRUS ENTRY MEDIATOR, ... | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacker, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2001-07-17 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
2IFF
 
 | |
3M1C
 
 | |
2GUM
 
 | |
2APR
 
 | |
1BQL
 
 | |
3HFM
 
 | |
1QS4
 
 | | Core domain of HIV-1 integrase complexed with Mg++ and 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-propenone | | Descriptor: | 1-(5-CHLOROINDOL-3-YL)-3-HYDROXY-3-(2H-TETRAZOL-5-YL)-PROPENONE, MAGNESIUM ION, PROTEIN (HIV-1 INTEGRASE (E.C.2.7.7.49)) | | Authors: | Goldgur, Y, Craigie, R, Fujiwara, T, Yoshinaga, T, Davies, D.R. | | Deposit date: | 1999-06-25 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1IOB
 
 | |
6APR
 
 | |
4APR
 
 | |
2FBJ
 
 | |
