1GKT
 
 | | Neutron Laue diffraction structure of endothiapepsin complexed with transition state analogue inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H261 | | Authors: | Coates, L, Erskine, P.T, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2001-08-20 | | Release date: | 2001-11-20 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | A Neutron Laue Diffraction Study of Endothiapepsin: Implications for the Aspartic Proteinase Mechanism
Biochemistry, 40, 2001
|
|
1GVW
 
 | | Endothiapepsin complex with PD-130,328 | | Descriptor: | ENDOTHIAPEPSIN, N-(tert-butoxycarbonyl)-L-phenylalanyl-N-{(1S)-1-[(R)-hydroxy(2-{[(2S)-2-methylbutyl]amino}-2-oxoethyl)phosphoryl]-3-methylbutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1GVV
 
 | | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes; implications for the Aspartic Proteinase Mechanism | | Descriptor: | ENDOTHIAPEPSIN, N-[(5S,9S,10S,13S)-9-hydroxy-5,10-bis(2-methylpropyl)-4,7,12,16-tetraoxo-3,6,11,17-tetraazabicyclo[17.3.1]tricosa-1(23),19,21-trien-13-yl]-3-(naphthalen-1-yl)-2-(naphthalen-1-ylmethyl)propanamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1GVT
 
 | | Endothiapepsin complex with CP-80,794 | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R,2S)-1-(cyclohexylmethyl)-2-hydroxy-3-(1-methylethoxy)-3-oxopropyl]-S-methyl-L-cysteinamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1GVU
 
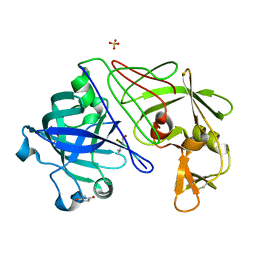 | | Endothiapepsin complex with H189 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR, H189, ... | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
2VS2
 
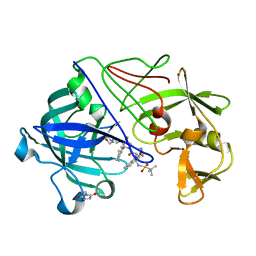 | | Neutron diffraction structure of endothiapepsin in complex with a gem- diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
5KMW
 
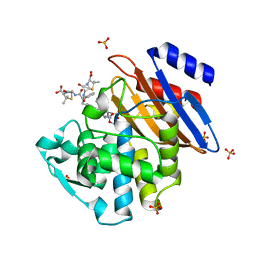 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
2JJJ
 
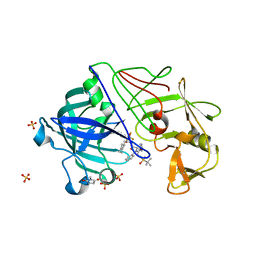 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
2JJI
 
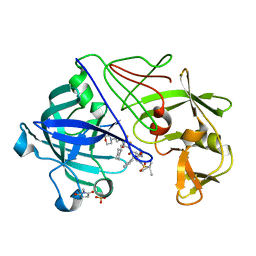 | | Endothiapepsin in complex with a gem-diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S.J, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-09 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
2C1H
 
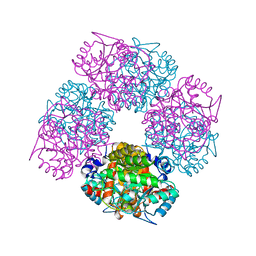 | | The X-ray Structure of Chlorobium vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor | | Descriptor: | 4,7-DIOXOSEBACIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S, Wood, S.P, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2005-09-14 | | Release date: | 2005-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Chlorobium Vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed with a Diacid Inhibitor.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1GVX
 
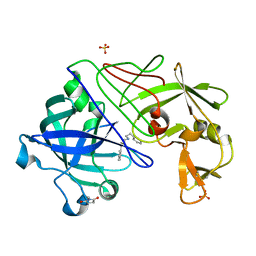 | | Endothiapepsin complexed with H256 | | Descriptor: | ENDOTHIAPEPSIN, INHIBITOR H256, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Crump, M.P, Wood, S.P, Cooper, J.B. | | Deposit date: | 2002-02-27 | | Release date: | 2002-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Five Atomic Resolution Structures of Endothiapepsin Inhibitor Complexes: Implications for the Aspartic Proteinase Mechanism
J.Mol.Biol., 318, 2002
|
|
1W1Z
 
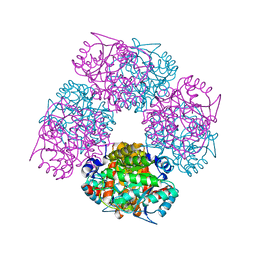 | | Structure of the plant like 5-Aminolaevulinic Acid Dehydratase from Chlorobium vibrioforme | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, LAEVULINIC ACID, MAGNESIUM ION | | Authors: | Coates, L, Beaven, G, Erskine, P.T, Beale, S.I, Avissar, Y.J, Gill, R, Mohammed, F, Wood, S.P, Shoolingin-Jordan, P, Cooper, J.B. | | Deposit date: | 2004-06-24 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The X-ray structure of the plant like 5-aminolaevulinic acid dehydratase from Chlorobium vibrioforme complexed with the inhibitor laevulinic acid at 2.6 A resolution.
J. Mol. Biol., 342, 2004
|
|
4C3Q
 
 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) at 100K | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1 | | Authors: | Coates, L, Tomanicek, S.J, Schrader, T, Weiss, K.L, Ng, J.D, Ostermann, A. | | Deposit date: | 2013-08-26 | | Release date: | 2014-07-09 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Cryogenic Neutron Protein Crystallography: Routine Methods and Potential Benefits
J.Appl.Crystallogr., 47, 2014
|
|
1OD1
 
 | | Endothiapepsin PD135,040 complex | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide, SULFATE ION | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Cooper, J.B. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-12 | | Last modified: | 2012-11-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The Structure of Endothiapepsin Complexed with the Gem-Diol Inhibitor Pd-135,040 at 1.37 A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
8E4J
 
 | |
8E4R
 
 | |
6C7A
 
 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C79
 
 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
1OEW
 
 | | ATOMIC RESOLUTION STRUCTURE OF NATIVE ENDOTHIAPEPSIN | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, SERINE, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
1OEX
 
 | | Atomic Resolution Structure of Endothiapepsin in Complex with a Hydroxyethylene Transition State Analogue Inhibitor H261 | | Descriptor: | ENDOTHIAPEPSIN, GLYCEROL, INHIBITOR H261, ... | | Authors: | Coates, L, Erskine, P.T, Mall, S, Gill, R.S, Wood, S.P, Myles, D.A.A, Cooper, J.B. | | Deposit date: | 2003-03-31 | | Release date: | 2003-04-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analysis of the Catalytic Site of an Aspartic Proteinase and an Unexpected Mode of Binding by Short Peptides
Protein Sci., 12, 2003
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6WQF
 
 | |
