3MMW
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
3MMU
 
 | | Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima | | Descriptor: | CADMIUM ION, Endoglucanase, NICKEL (II) ION | | Authors: | Pereira, J.H, Chen, Z, McAndrew, R.P, Sapra, R, Chhabra, S.R, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2010-04-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima.
J.Struct.Biol., 172, 2010
|
|
4JVD
 
 | |
4JVI
 
 | |
4JVC
 
 | | Crystal structure of PqsR co-inducer binding domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Transcriptional regulator MvfR | | Authors: | Ilangovan, A, Emsley, J, Williams, P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR).
Plos Pathog., 9, 2013
|
|
4NBU
 
 | | Crystal structure of FabG from Bacillus sp | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase, ACETOACETYL-COENZYME A | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBT
 
 | | Crystal structure of FabG from Acholeplasma laidlawii | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBV
 
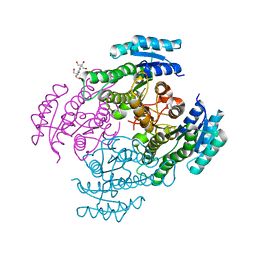 | | Crystal structure of FabG from Cupriavidus taiwanensis | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-[acyl-carrier-protein] reductase putative short-chain dehydrogenases/reductases (SDR) family protein | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBW
 
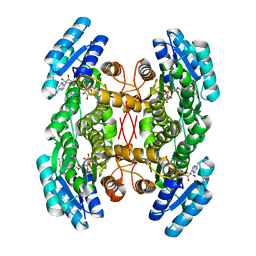 | | Crystal structure of FabG from Plesiocystis pacifica | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
1NX4
 
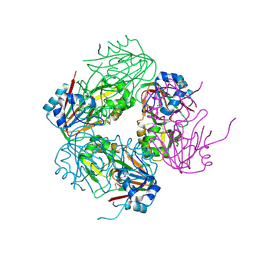 | | The crystal structure of carbapenem synthase (CarC) | | Descriptor: | 2-OXOGLUTARIC ACID, Carbapenem synthase, FE (III) ION | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-08 | | Release date: | 2003-06-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
1NX8
 
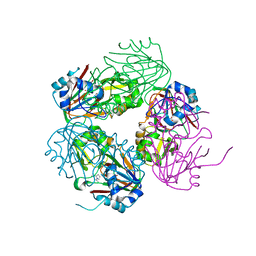 | | Structure of carbapenem synthase (CarC) complexed with N-acetyl proline | | Descriptor: | 1-ACETYL-L-PROLINE, 2-OXOGLUTARIC ACID, Carbapenem synthase, ... | | Authors: | Clifton, I.J, Doan, L.X, Sleeman, M.C, Topf, M, Suzuki, H, Wilmouth, R.C, Schofield, C.J. | | Deposit date: | 2003-02-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of carbapenem synthase (CarC).
J.Biol.Chem., 278, 2003
|
|
