7PJY
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in chimeric state 1 (CHI1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJX
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJZ
 
 | | Structure of the 70S-EF-G-GDP ribosome complex with tRNAs in chimeric state 2 (CHI2-EF-G-GDP) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJW
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 2 (H2-EF-G-GDP-Pi) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJS
 
 | | Structure of the 70S ribosome with tRNAs in the classical pre-translocation state and apramycin (C) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement
Nat Commun, 12, 2021
|
|
7PJV
 
 | | Structure of the 70S-EF-G-GDP-Pi ribosome complex with tRNAs in hybrid state 1 (H1-EF-G-GDP-Pi) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJT
 
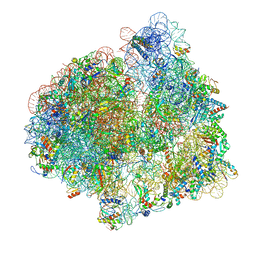 | | Structure of the 70S ribosome with tRNAs in hybrid state 1 (H1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-10-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
7PJU
 
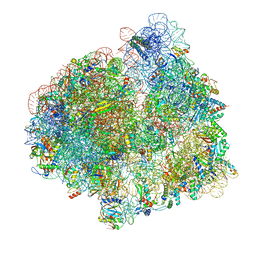 | | Structure of the 70S ribosome with tRNAs in hybrid state 2 (H2) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petrychenko, V, Peng, B.Z, Schwarzer, A.C, Peske, F, Rodnina, M.V, Fischer, N. | | Deposit date: | 2021-08-24 | | Release date: | 2021-11-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structural mechanism of GTPase-powered ribosome-tRNA movement.
Nat Commun, 12, 2021
|
|
4PXK
 
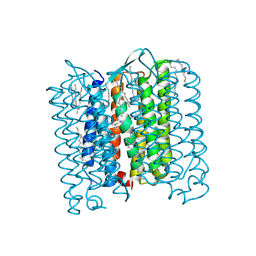 | | Crystal structure of Haloarcula marismortui bacteriorhodopsin I D94N mutant | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Shevchenko, V, Gushchin, I, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2014-03-24 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Escherichia coli-Expressed Haloarcula marismortui Bacteriorhodopsin I in the Trimeric Form.
Plos One, 9, 2014
|
|
8RG0
 
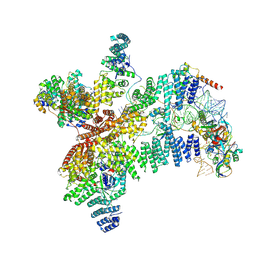 | | Structure of human eIF3 core from closed 48S translation initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ6
 
 | | Structure of human 48S translation initiation complex with initiator tRNA, eIF1A and eIF3 (off-pathway) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ2
 
 | | Structure of human 48S translation initiation complex in AUG recognition state after eIF5-induced GTP hydrolysis by eIF2 (48S-2) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ4
 
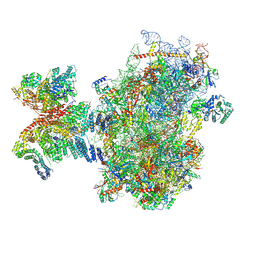 | | Structure of human 48S translation initiation complex after eIF5 release (48S-4) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ5
 
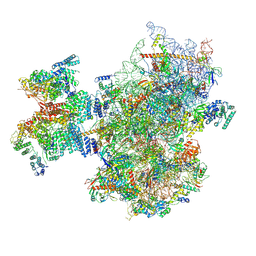 | | Structure of human 48S translation initiation complex after eIF2 release prior 60S subunit joining (48S-5) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ3
 
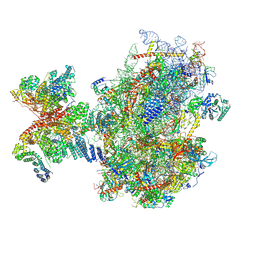 | | Structure of human 48S translation initiation complex upon transfer of initiator tRNA to eIF5B (48S-3) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
8PJ1
 
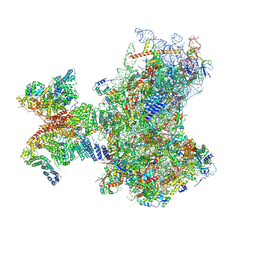 | | Structure of human 48S translation initiation complex in open codon scanning state (48S-1) | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-06-22 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex from codon scanning toward subunit joining
Nat.Struct.Mol.Biol., 2024
|
|
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
7AVN
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
7AVP
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in violet form | | Descriptor: | Bacteriorhodopsin, EICOSANE | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
7AVO
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1211 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
6W25
 
 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
4XTO
 
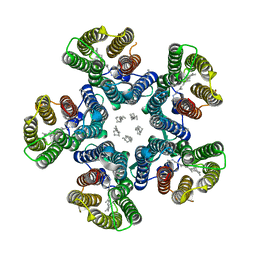 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTN
 
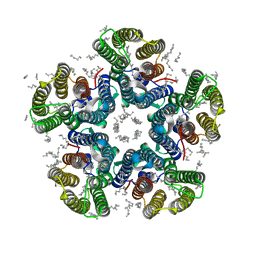 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 4.9 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XTL
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric blue form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, SODIUM ION, ... | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6Y83
 
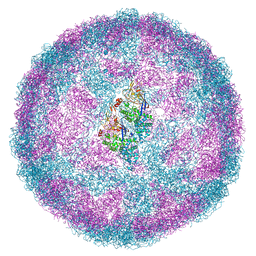 | | Capsid structure of Leishmania RNA virus 1 | | Descriptor: | Capsid protein | | Authors: | Prochazkova, M, Fuzik, T, Grybtchuk, D, Falginella, F, Podesvova, L, Yurchenko, V, Vacha, R, Plevka, P. | | Deposit date: | 2020-03-03 | | Release date: | 2020-11-11 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Capsid Structure of Leishmania RNA Virus 1.
J.Virol., 95, 2021
|
|
