6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6Y6X
 
 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
8K82
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, Not5 and P-site tRNA | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8K2D
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7P3K
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide (control) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Buschauer, R, Komar, T, Becker, T, Berninghausen, O, Cheng, J, Beckmann, R. | | Deposit date: | 2021-07-08 | | Release date: | 2021-10-27 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
6Q8Y
 
 | | Cryo-EM structure of the mRNA translating and degrading yeast 80S ribosome-Xrn1 nuclease complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Heckel, E, Cheng, J, Buschauer, R, Kater, L, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2018-12-16 | | Release date: | 2019-03-13 | | Last modified: | 2025-12-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the 80S ribosome-Xrn1 nuclease complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7ZW0
 
 | | FAP-80S Complex - Rotated state | | Descriptor: | 18S ribosomal RNA (RDN18-1), 25S ribosomal RNA (RDN25-1), 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover.
Mol.Cell, 82, 2022
|
|
5M5H
 
 | | RIBOSOME-BOUND YIDC INSERTASE | | Descriptor: | Membrane protein insertase YidC | | Authors: | Kedrov, A, Wickles, S, Crevenna, A.H, van der Sluis, E, Buschauer, R, Berninghausen, O, Lamb, D.C, Beckmann, R. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis.
Cell Rep, 17, 2016
|
|
6T83
 
 | | Structure of yeast disome (di-ribosome) stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7T
 
 | | Structure of yeast 80S ribosome stalled on poly(A) tract. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-23 | | Release date: | 2019-12-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T7I
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6T4Q
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CCG inhibitory codon combination. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-14 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6SV4
 
 | | The cryo-EM structure of SDD1-stalled collided trisome. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-09-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SNT
 
 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2025-12-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QGH
 
 | | Structure of the E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QH4
 
 | | Structure of the B. subtilis disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.45 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QG8
 
 | | Structure of the collided E. coli disome - VemP-stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-07 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGU
 
 | | Structure of the B. subtilis disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGN
 
 | | Structure of the SmrB-bound E. coli disome - stalled 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-09 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7QGR
 
 | | Structure of the SmrB-bound E. coli disome - collided 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Kratzat, H, Buschauer, R, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome collisions induce mRNA cleavage and ribosome rescue in bacteria.
Nature, 603, 2022
|
|
7OZS
 
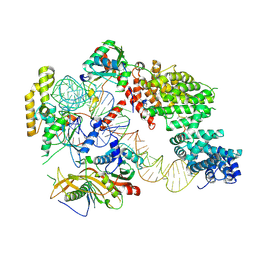 | | Structure of the hexameric 5S RNP from C. thermophilum | | Descriptor: | 5S rRNA, 60S ribosomal protein l5-like protein, Putative ribosomal protein, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2021-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 2023
|
|
8AGT
 
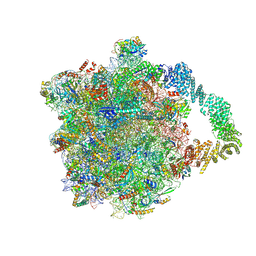 | | Yeast RQC complex in state F | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AAF
 
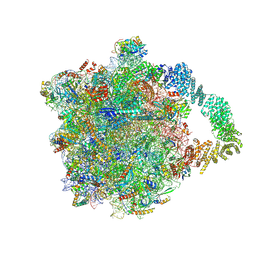 | | Yeast RQC complex in state G | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
8AGU
 
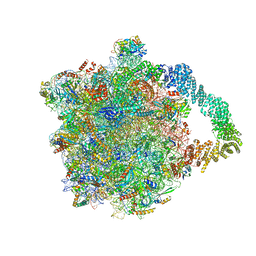 | | Yeast RQC complex in state E | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
