6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
5JF4
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT019 | | Descriptor: | (3R)-3-{3-[(1-benzofuran-3-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF5
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT020 | | Descriptor: | (3R)-3-{3-[(2H-1,3-benzodioxol-5-yl)methyl]-1,2,4-oxadiazol-5-yl}-4-cyclopentyl-N-hydroxybutanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5N9I
 
 | |
5N9H
 
 | |
5NXN
 
 | |
5NXR
 
 | |
5NXU
 
 | |
4D65
 
 | |
4D64
 
 | |
5JEZ
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Ser | | Descriptor: | ACETATE ION, Met-Ala-Ser, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF7
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor SMP289 | | Descriptor: | 2-(3-benzyl-5-bromo-1H-indol-1-yl)-N-hydroxyacetamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF1
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with actinonin | | Descriptor: | ACETATE ION, ACTINONIN, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5LDT
 
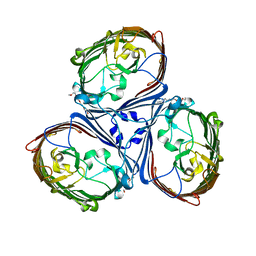 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CALCIUM ION, MOMP porin | | Authors: | Ferrara, L.G.M, Wallat, G.D, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-27 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
5JF0
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with tripeptide Met-Ala-Arg | | Descriptor: | ACETATE ION, MET-ALA-ARG, NICKEL (II) ION, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF2
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT002 | | Descriptor: | (3R)-3-{3-[(4-fluorophenyl)methyl]-1,2,4-oxadiazol-5-yl}-N-hydroxyheptanamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JEY
 
 | |
5JF6
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor 6b (AB47) | | Descriptor: | 2-(5-bromo-1H-indol-3-yl)-N-hydroxyacetamide, ACETATE ION, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T, Hamiche, K. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF8
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor RAS358 (21) | | Descriptor: | ACETATE ION, IMIDAZOLE, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JEX
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae, crystallized in imidazole buffer | | Descriptor: | IMIDAZOLE, Peptide deformylase, ZINC ION | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5JF3
 
 | | Crystal structure of type 2 PDF from Streptococcus agalactiae in complex with inhibitor AT018 | | Descriptor: | ACETATE ION, IMIDAZOLE, Peptide deformylase, ... | | Authors: | Fieulaine, S, Giglione, C, Meinnel, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A unique peptide deformylase platform to rationally design and challenge novel active compounds.
Sci Rep, 6, 2016
|
|
5LDV
 
 | | Crystal Structures of MOMP from Campylobacter jejuni | | Descriptor: | CALCIUM ION, MOMP porin, N-OCTANE, ... | | Authors: | Wallat, G.D, Ferrara, L.M.G, Moynie, L, Naismith, J.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone.
J.Mol.Biol., 428, 2016
|
|
