8BAW
 
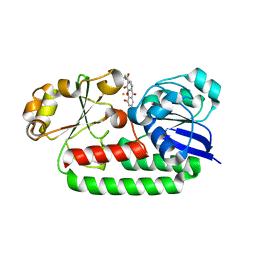 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - 5-LICAM siderophore analogue complex. | | Descriptor: | FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Booth, R, Dodson, E.J, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BJ9
 
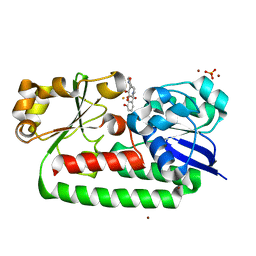 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - 5LICAM complex. | | Descriptor: | ABC transporter, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide), ... | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BNW
 
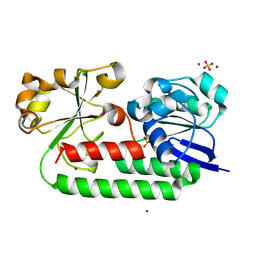 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - apo form | | Descriptor: | ABC transporter, NICKEL (II) ION, SULFATE ION | | Authors: | Blagova, E.V, Bennett, M, Booth, R, Dodson, E.J, Duhme-KLair, A.-K, Wilson, K.S. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BAX
 
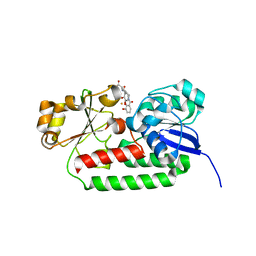 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - azotochelin complex. | | Descriptor: | Azotochelin, FE (III) ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Blagova, E.V, Miller, A, Dodson, E.J, Booth, R, Duhme-Klair, A.K, Wilson, K.S. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8BF6
 
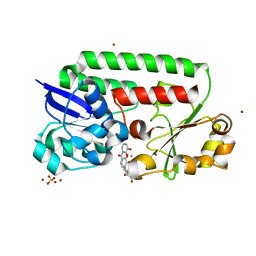 | | X-ray structure of the CeuE Homologue from Parageobacillus thermoglucosidasius - azotochelin complex | | Descriptor: | ABC transporter, Azotochelin, FE (III) ION, ... | | Authors: | Wilson, K.S, Duhme-Klair, A.-K, Blagova, E.V, Miller, A, Booth, R, Dodson, E.J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B7X
 
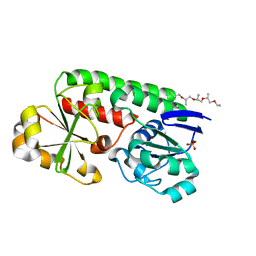 | | X-ray structure of the CeuE Homologue from Geobacillus stearothermophilus - apo form. | | Descriptor: | O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), SULFATE ION, Siderophore ABC transporter substrate-binding protein | | Authors: | Wilson, K.S, Duhme-Klair, A.K, Blagova, E.V, Bennett, M. | | Deposit date: | 2022-10-03 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thermostable homologues of the periplasmic siderophore-binding protein CeuE from Geobacillus stearothermophilus and Parageobacillus thermoglucosidasius.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1T9H
 
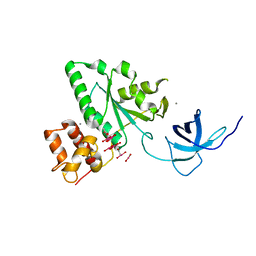 | | The crystal structure of YloQ, a circularly permuted GTPase. | | Descriptor: | ACETATE ION, CALCIUM ION, Probable GTPase engC, ... | | Authors: | Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Cladiere, L, Antson, A.A, Isupov, M.N, Seror, S.J, Wilkinson, A.J. | | Deposit date: | 2004-05-17 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of YloQ, a Circularly Permuted GTPase Essential for Bacillus Subtilis Viability.
J.Mol.Biol., 340, 2004
|
|
1K3F
 
 | | Uridine Phosphorylase from E. coli, Refined in the Monoclinic Crystal Lattice | | Descriptor: | uridine phosphorylase | | Authors: | Morgunova, E.Yu, Mikhailov, A.M, Popov, A.N, Blagova, E.V, Smirnova, E.A, Vainshtein, B.K, Mao, C, Armstrong, S.R, Ealick, S.E, Komissarov, A.A, Linkova, E.V, Burlakova, A.A, Mironov, A.S, Debabov, V.G. | | Deposit date: | 2001-10-02 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic structure at 2.5 A resolution of uridine phosphorylase from E. coli as refined in the monoclinic crystal lattice.
FEBS Lett., 367, 1995
|
|
5M1N
 
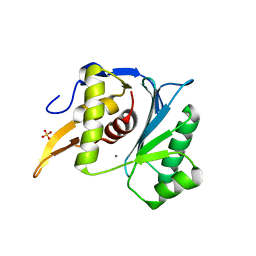 | | Crystal structure of the large terminase nuclease from thermophilic phage G20c with bound Manganese | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, Phage terminase large subunit, ... | | Authors: | Xu, R.G, Jenkins, H.T, Chechik, M, Blagova, E.V, Greive, S.J, Antson, A.A. | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Viral genome packaging terminase cleaves DNA using the canonical RuvC-like two-metal catalysis mechanism.
Nucleic Acids Res., 45, 2017
|
|
5MBU
 
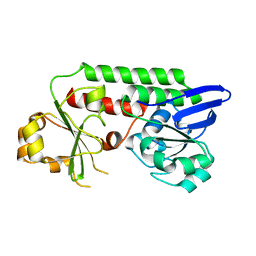 | | CeuE (H227A, Y288F variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5MBQ
 
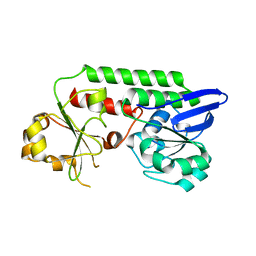 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
2GX5
 
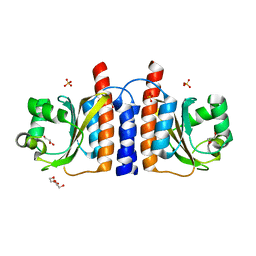 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, GTP-sensing transcriptional pleiotropic repressor codY, ... | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria.
J.Biol.Chem., 281, 2006
|
|
1P3E
 
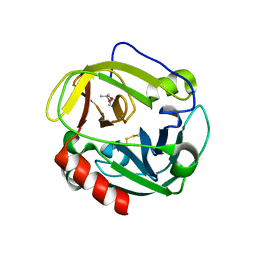 | | Structure of Glu endopeptidase in complex with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
1P3C
 
 | | Glutamyl endopeptidase from Bacillus intermedius | | Descriptor: | glutamyl-endopeptidase | | Authors: | Meijers, R, Blagova, E.V, Levdikov, V.M, Rudenskaya, G.N, Chestukhina, G.G, Akimkina, T.V, Kostrov, S.V, Lamzin, V.S, Kuranova, I.P. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of glutamyl endopeptidase from Bacillus intermedius reveals a structural link between zymogen activation and charge compensation.
Biochemistry, 43, 2004
|
|
2HGV
 
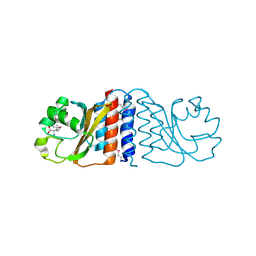 | |
3DD6
 
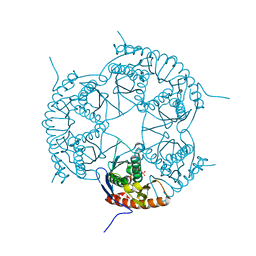 | | Crystal structure of Rph, an exoribonuclease from Bacillus anthracis at 1.7 A resolution | | Descriptor: | Ribonuclease PH, SULFATE ION | | Authors: | Rawlings, A.E, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Wilson, K.S, Wilkinson, A.J, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2008-06-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | The structure of Rph, an exoribonuclease from Bacillus anthracis, at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2PFM
 
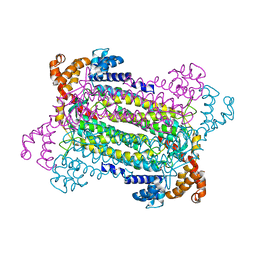 | | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis | | Descriptor: | Adenylosuccinate lyase, MALONATE ION | | Authors: | Levdikov, V.M, Blagova, E.V, Baumgart, M, Moroz, O.V, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2007-04-05 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase (PurB) from Bacillus anthracis
To be Published
|
|
2WCF
 
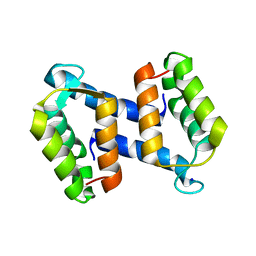 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WC8
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | CITRIC ACID, PROTEIN S100-A12, SODIUM ION, ... | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCE
 
 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
2WCB
 
 | | S100A12 complex with zinc in the absence of calcium | | Descriptor: | PROTEIN S100-A12, SODIUM ION, ZINC ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
5MBT
 
 | | CeuE (H227L, Y288F variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
4CSB
 
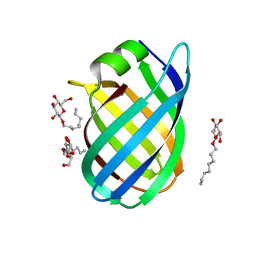 | | Structure of the Virulence-Associated Protein VapD from the intracellular pathogen Rhodococcus equi. | | Descriptor: | VIRULENCE ASSOCIATED PROTEIN VAPD, octyl beta-D-glucopyranoside | | Authors: | Whittingham, J.L, Blagova, E.V, Finn, C.E, Luo, H, Miranda-CasoLuengo, R, Turkenburg, J.P, Leech, A.P, Walton, P.H, Meijers, W.G, Wilkinson, A.J. | | Deposit date: | 2014-03-06 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Virulence-Associated Protein Vapd from the Intracellular Pathogen Rhodococcus Equi.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3O9P
 
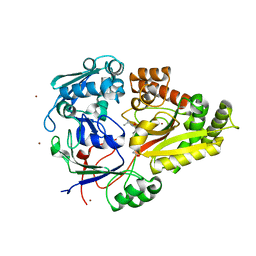 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
3TUF
 
 | |
