4QAJ
 
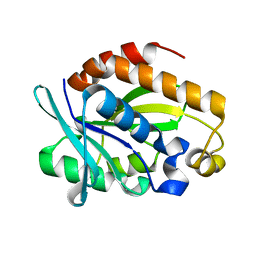 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
4QBK
 
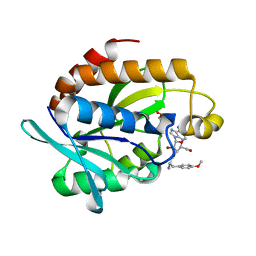 | | Crystal structure of the complex of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with amino acyl-tRNA analogue at 1.77 Angstrom resolution | | Descriptor: | 3'-deoxy-3'-[(O-methyl-L-tyrosyl)amino]adenosine, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-08 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
1TGM
 
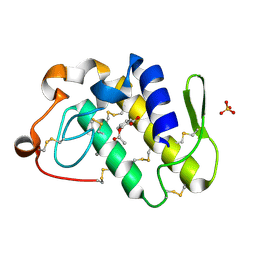 | | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution
To be Published
|
|
4Q22
 
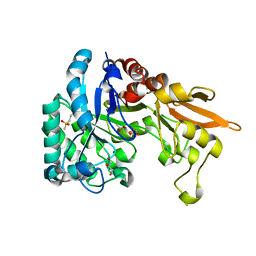 | | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-04-05 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine at 1.93 Angstrom resolution
To be Published
|
|
4QT4
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Angstrom resolution shows the Closed Structure of the Substrate Binding Cleft | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 angstrom resolution shows the closed structure of the substrate-binding cleft.
FEBS Open Bio, 4, 2014
|
|
1N76
 
 | | CRYSTAL STRUCTURE OF HUMAN SEMINAL LACTOFERRIN AT 3.4 A RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN, ... | | Authors: | Kumar, J, Weber, W, Munchau, S, Yadav, S, Singh, S.B, Sarvanan, K, Paramsivam, M, Sharma, S, Kaur, P, Bhushan, A, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of human seminal lactoferrin at 3.4A resolution
Indian J.Biochem.Biophys., 40, 2003
|
|
7DLQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF LACTOPEROXIDASE WITH HYDROGEN PEROXIDE AT 1.77A RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2020-11-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
2Z5Z
 
 | | Crystal structure of the complex of buffalo Lactoperoxidase with fluoride ion at 3.5A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sheikh, I.A, Jain, R, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the complex of buffalo Lactoperoxidase with fluoride ion at 3.5A resolution
To be Published
|
|
4Q8S
 
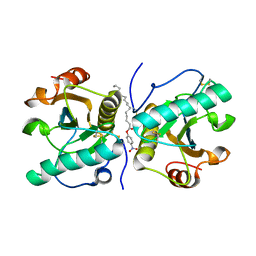 | | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrophenyl hexadecanoate, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of mammalian Peptidoglycan recognition protein PGRP-S with paranitrophenyl palmitate and N-acetyl glucosamine at 2.09 A resolution
To be Published
|
|
1OWQ
 
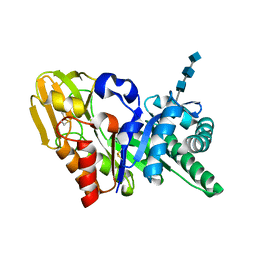 | | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, signal processing protein | | Authors: | Kumar, J, Sharma, S, Jasti, J, Bhushan, A, Singh, T.P. | | Deposit date: | 2003-03-29 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 40 kDa signalling protein (SPC-40) secreted during involution
To be Published
|
|
3OSZ
 
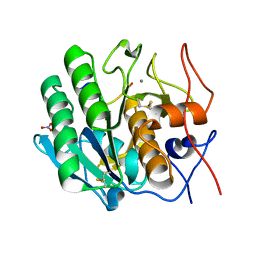 | | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution | | Descriptor: | 10-mer peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Singh, A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-09-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the complex of proteinase K with an antimicrobial nonapeptide, at 2.26 A resolution
To be Published
|
|
6LF7
 
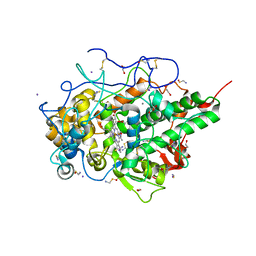 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Tyagi, T.K, Singh, R.P, Singh, A.K, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-30 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution.
To Be Published
|
|
3TUS
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXYBENZOIC ACID, ... | | Authors: | Shukla, P.K, Gautam, L, Singh, A, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution
To be Published
|
|
4JC4
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 2.25 angstrom resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2013-02-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
3M7S
 
 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
3CG9
 
 | | Crystal structure of the complex of peptidoglycan recognition protein with methyloxane-2,3,4,5-tetrol at 2.9 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-L-rhamnopyranose | | Authors: | Sharma, P, Kaur, A, Singh, N, Sharma, S, Bhushan, A, Pathak, K.M.L, Kaur, P, Singh, T.P. | | Deposit date: | 2008-03-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with methyoxane-2,3,4,5-tetrol at 2.9 A resolution
To be Published
|
|
3D5H
 
 | | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION | | Authors: | Kumar, S, Singh, N, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-05-16 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of haementhin from Haemanthus multiflorus at 2.0A resolution: Formation of a novel loop on a TIM barrel fold and its functional significance
To be Published
|
|
3ERI
 
 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sheikh, I.A, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3ERH
 
 | | First structural evidence of substrate specificity in mammalian peroxidases: Crystal structures of substrate complexes with lactoperoxidases from two different species | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sinha, M, Singh, S.B, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-10-02 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3FG5
 
 | | Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution | | Descriptor: | AJMALINE, Group II Phospholipase A2, pentapeptide FLSYK | | Authors: | Kumar, M, Kumar, S, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2008-12-05 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution
To be Published
|
|
2FA7
 
 | | Crystal structure of the complex of bovine lactoferrin C-lobe with a pentasaccharide at 2.38 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, N, Jain, R, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2005-12-07 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of the complex of bovine lactoferrin C-lobe with a pentasaccharide at 2.38 A resolution
To be Published
|
|
2HPZ
 
 | | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution | | Descriptor: | 11-mer synthetic peptide, CALCIUM ION, NITRATE ION, ... | | Authors: | Prem kumar, R, Singh, A.K, Somvanshi, R.K, Singh, N, Sharma, S, Kaur, P, Dey, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-18 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of proteinase K complex with a synthetic peptide KLKLLVVIRLK at 1.69 A resolution
To be Published
|
|
4GUW
 
 | | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of type 1 Ribosome inactivating protein from Momordica balsamina with lipopolysaccharide at 1.6 Angstrom resolution
To be published
|
|
4H0Z
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl muramic acid at 2.0 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-muramic acid, rRNA N-glycosidase | | Authors: | Singh, A, Pandey, S, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with N-acetyl muramic acid at 2.0 Angstrom resolution
TO BE PUBLISHED
|
|
4HSD
 
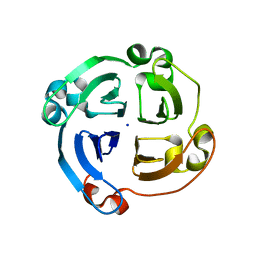 | | Crystal structure of a new form of plant lectin from Cicer arietinum at 2.45 Angstrom resolution | | Descriptor: | Lectin, SODIUM ION | | Authors: | Kumar, S, Singh, A, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a new form of plant lectin from Cicer arietinum at 2.45 Angstrom resolution
to be published
|
|
