4UBP
 
 | | STRUCTURE OF BACILLUS PASTEURII UREASE INHIBITED WITH ACETOHYDROXAMIC ACID AT 1.55 A RESOLUTION | | Descriptor: | ACETOHYDROXAMIC ACID, NICKEL (II) ION, PROTEIN (UREASE (CHAIN A)), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1999-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The complex of Bacillus pasteurii urease with acetohydroxamate anion from X-ray data at 1.55 A resolution.
J.Biol.Inorg.Chem., 5, 2000
|
|
2UBP
 
 | | STRUCTURE OF NATIVE UREASE FROM BACILLUS PASTEURII | | Descriptor: | NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), PROTEIN (UREASE BETA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-11-04 | | Release date: | 1999-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
3UBP
 
 | | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | | Descriptor: | DIAMIDOPHOSPHATE, NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Miletti, S, Mangani, S, Ciurli, S. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
1C75
 
 | | 0.97 A "AB INITIO" CRYSTAL STRUCTURE OF CYTOCHROME C-553 FROM BACILLUS PASTEURII | | Descriptor: | CYTOCHROME C-553, HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2000-02-09 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
1S3T
 
 | | BORATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | BORIC ACID, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-04-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism.
J.Am.Chem.Soc., 126, 2004
|
|
1UBP
 
 | | CRYSTAL STRUCTURE OF UREASE FROM BACILLUS PASTEURII INHIBITED WITH BETA-MERCAPTOETHANOL AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | BETA-MERCAPTOETHANOL, NICKEL (II) ION, UREASE | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 1998-01-21 | | Release date: | 1999-03-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The complex of Bacillus pasteurii urease with beta-mercaptoethanol from X-ray data at 1.65-A resolution
J.Biol.Inorg.Chem., 3, 1998
|
|
4CEX
 
 | | 1.59 A resolution Fluoride inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, FLUORIDE ION, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Fluoride Inhibition of Sporosarcina Pasteurii Urease: Structure and Thermodynamics.
J.Biol.Inorg.Chem., 19, 2014
|
|
4CEU
 
 | | 1.58 A resolution native Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fluoride Inhibition of Sporosarcina Pasteurii Urease: Structure and Thermodynamics.
J.Biol.Inorg.Chem., 19, 2014
|
|
4AC7
 
 | | The crystal structure of Sporosarcina pasteurii urease in complex with citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, HYDROXIDE ION, ... | | Authors: | Benini, S, Kosikowska, P, Cianci, M, Gonzalez Vara, A, Berlicki, L, Ciurli, S. | | Deposit date: | 2011-12-14 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of Sporosarcina Pasteurii Urease in a Complex with Citrate Provides New Hints for Inhibitor Design.
J.Biol.Inorg.Chem., 18, 2013
|
|
2J9B
 
 | | THE CRYSTAL STRUCTURE OF CYTOCHROME C' FROM RUBRIVIVAX GELATINOSUS AT 1.5 A RESOLUTION AND PH 6.3 | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2006-11-06 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structure of Rubrivivax gelatinosus cytochrome c'.
J. Inorg. Biochem., 102, 2008
|
|
2Y3Y
 
 | | Holo-Ni(II) HpNikR is a symmetric tetramer containing four canonic square-planar Ni(II) ions at physiological pH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, NICKEL (II) ION, ... | | Authors: | Benini, S, Cianci, M, Ciurli, S. | | Deposit date: | 2011-01-04 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Holo-Ni(2+)Helicobacter Pylori Nikr Contains Four Square-Planar Nickel-Binding Sites at Physiological Ph.
Dalton Trans, 40, 2011
|
|
2CMP
 
 | | crystal structure of the DNA binding domain of G1P SMALL TERMINASE SUBUNIT from bacteriophage SF6 | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | Benini, S, Chechik, M, Ortiz-Lombardia, M, Polier, S, Shevtsov, M.B, DeLuchi, D, Alonso, J.C, Antson, A.A. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The 1.58 A Resolution Structure of the DNA-Binding Domain of Bacteriophage Sf6 Small Terminase Provides New Hints on DNA Binding
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1WCF
 
 | |
2J8W
 
 | | The crystal structure of cytochrome c' from Rubrivivax gelatinosus at 1.3 A Resolution and pH 8.0 | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High resolution crystal structure of Rubrivivax gelatinosus cytochrome c'.
J. Inorg. Biochem., 102, 2008
|
|
1SXV
 
 | |
1RFE
 
 | | Crystal structure of conserved hypothetical protein Rv2991 from Mycobacterium tuberculosis | | Descriptor: | hypothetical protein Rv2991 | | Authors: | Benini, S, Haouz, A, Proux, F, Betton, J.M, Alzari, P, Dodson, G.G, Wilson, K.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-28 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Rv2991 from Mycobacterium tuberculosis: An F420binding protein with unknown function.
J. Struct. Biol., 2019
|
|
1B7V
 
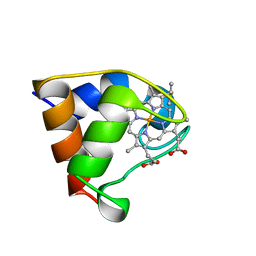 | | Structure of the C-553 cytochrome from Bacillus pasteruii to 1.7 A resolution | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C-553) | | Authors: | Gonzalez, A, Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S. | | Deposit date: | 1999-01-22 | | Release date: | 2000-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-A resolution.
Biochemistry, 39, 2000
|
|
4D74
 
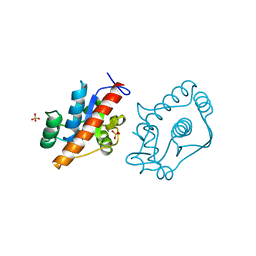 | | 1.57 A crystal structure of erwinia amylovora tyrosine phosphatase amsI | | Descriptor: | PROTEIN-TYROSINE-PHOSPHATASE AMSI, SULFATE ION | | Authors: | Benini, S, Salomone-Stagni, M, Caputi, L, Cianci, M. | | Deposit date: | 2014-11-19 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Characterization and 1.57 A Resolution Structure of the Key Fire Blight Phosphatase Amsi from Erwinia Amylovora
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4D48
 
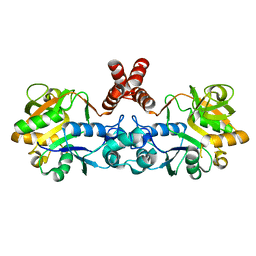 | | Crystal Structure of glucose-1-phosphate uridylyltransferase GalU from Erwinia amylovora. | | Descriptor: | GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Toccafondi, M, Wuerges, J, Cianci, M, Benini, S. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Glucose-1-phosphate uridylyltransferase from Erwinia amylovora: Activity, structure and substrate specificity.
Biochim. Biophys. Acta, 1865, 2017
|
|
1HLQ
 
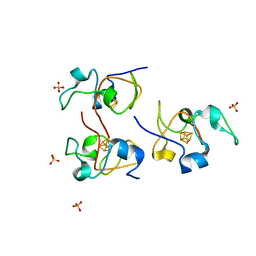 | | CRYSTAL STRUCTURE OF RHODOFERAX FERMENTANS HIGH POTENTIAL IRON-SULFUR PROTEIN REFINED TO 1.45 A | | Descriptor: | HIGH-POTENTIAL IRON-SULFUR PROTEIN, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Gonzalez, A, Ciurli, S, Benini, S. | | Deposit date: | 2000-12-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Rhodoferax fermentans high-potential iron-sulfur protein solved by MAD.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7OSO
 
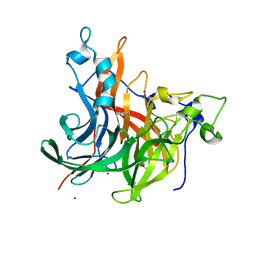 | | The crystal structure of Erwinia tasmaniensis levansucrase in complex with (S)-1,2,4-butanentriol | | Descriptor: | (2~{S})-butane-1,2,4-triol, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | Authors: | Polsinelli, I, Salomone-Stagni, M, Benini, S. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Erwinia tasmaniensis levansucrase shows enantiomer selection for (S)-1,2,4-butanetriol.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
5FRK
 
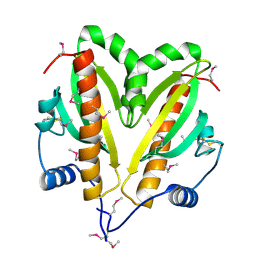 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-18 | | Release date: | 2017-02-15 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
5FR7
 
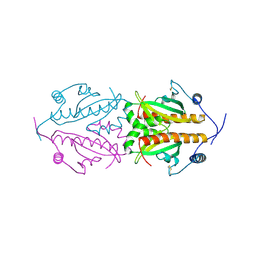 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
8KD8
 
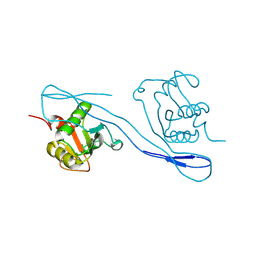 | |
