1E0G
 
 | | LYSM Domain from E.coli MLTD | | Descriptor: | Membrane-bound lytic murein transglycosylase D | | Authors: | Bateman, A, Bycroft, M. | | Deposit date: | 2000-03-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of a Lysm Domain from E.Coli Membrane Bound Lytic Murein Transglycosylase D (Mltd)
J.Mol.Biol., 299, 2000
|
|
5NB9
 
 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NBB
 
 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
6S5Z
 
 | | Structure of Rib R28N from Streptococcus pyogenes | | Descriptor: | SODIUM ION, Surface protein R28 | | Authors: | Whelan, F, Griffiths, S.C, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5W
 
 | | Structure of Rib domain 'Rib Long' from Lactobacillus acidophilus | | Descriptor: | SODIUM ION, SULFATE ION, Surface protein | | Authors: | Griffiths, S.C, Cooper, R.E.M, Whelan, F, Whittingham, J.L, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SX1
 
 | | Structure of Rib Standard, a Rib domain from Lactobacillus acidophilus | | Descriptor: | Surface protein | | Authors: | Cooper, R.E.M, Griffiths, S.C, Turkenburg, J.P, Bateman, A, Potts, J.R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5X
 
 | | Structure of RibR, the most N-terminal Rib domain from Group B Streptococcus species Streptococcus agalactiae | | Descriptor: | Group B streptococcal R4 surface protein, SODIUM ION | | Authors: | Whelan, F, Turkenburg, J.P, Griffiths, S.C, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6S5Y
 
 | |
3DCX
 
 | |
8R0N
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 10.5 in detergent in the ground state | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0L
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 8.0 in nanodisc | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0O
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 10.5 in detergent in the M state | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0M
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 8.0 in detergent | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0K
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR1 at pH 4.3 in detergent | | Descriptor: | EICOSANE, RETINAL, Rhodopsin | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8R0P
 
 | | Cryo-EM structure of the microbial rhodopsin CryoR2 at pH 8.0 in detergent | | Descriptor: | EICOSANE, RETINAL, microbial rhodopsin CryoR2 | | Authors: | Kovalev, K, Marin, E, Stetsenko, A, Guskov, A, Lamm, G.H.U. | | Deposit date: | 2023-10-31 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | CryoRhodopsins: a new clade of microbial rhodopsins from cold environments
To Be Published
|
|
8C3T
 
 | |
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
3HSA
 
 | |
3H0N
 
 | |
6CKU
 
 | | Solution structure of the zebrafish granulin AaE | | Descriptor: | Granulin-AaE | | Authors: | Wang, P, Ni, F. | | Deposit date: | 2018-02-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of zebrafish progranulins identifies a well-folded granulin/epithelin module protein with pro-cell survival activities.
Protein Sci., 27, 2018
|
|
2FU2
 
 | | Crystal structure of protein SPy2152 from Streptococcus pyogenes | | Descriptor: | Hypothetical protein SPy2152 | | Authors: | Chang, C, Cymborowski, M, Otwinowski, Z, Minor, W, Lezondra, L.-E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-25 | | Release date: | 2006-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of pyogenecin immunity protein, a novel bacteriocin-like immunity protein from Streptococcus pyogenes.
Bmc Struct.Biol., 9, 2009
|
|
8QQZ
 
 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QR0
 
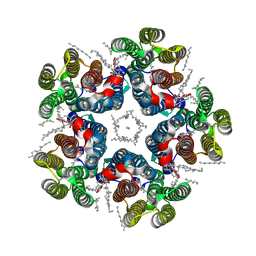 | | Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3 | | Descriptor: | Bacteriorhodopsin-like protein, DODECYL-BETA-D-MALTOSIDE, EICOSANE | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Marin, E, Stetsenko, A, Guskov, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLE
 
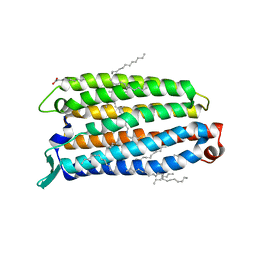 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 4.6 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
8QLF
 
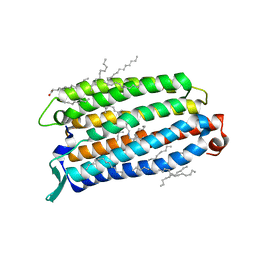 | | Crystal structure of the light-driven sodium pump ErNaR in the monomeric form at pH 8.8 | | Descriptor: | Bacteriorhodopsin-like protein, EICOSANE, OLEIC ACID | | Authors: | Kovalev, K, Podoliak, E, Lamm, G.H.U, Astashkin, R, Bourenkov, G. | | Deposit date: | 2023-09-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A subgroup of light-driven sodium pumps with an additional Schiff base counterion.
Nat Commun, 15, 2024
|
|
