3BTN
 
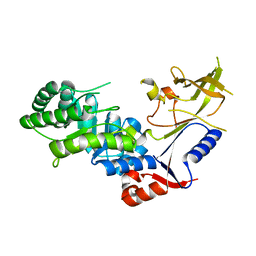 | | Crystal structure of antizyme inhibitor, an ornithine decarboxylase homologous protein | | Descriptor: | Antizyme inhibitor 1 | | Authors: | Dym, O, Unger, T, Albeck, S, Kahana, C, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function.
Protein Sci., 17, 2008
|
|
6FHE
 
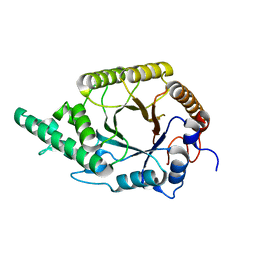 | | Highly active enzymes by automated modular backbone assembly and sequence design | | Descriptor: | Synthetic construct | | Authors: | Lapidot, G, Khersonsky, O, Lipsh, R, Dym, O, Albeck, S, Rogotner, S, Fleishman, J.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Highly active enzymes by automated combinatorial backbone assembly and sequence design.
Nat Commun, 9, 2018
|
|
7O4D
 
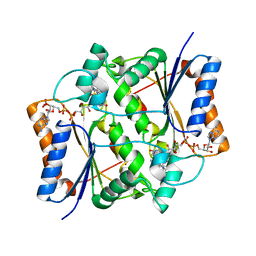 | | QR2 inhibitor from a novel sulfanamide series to tackle age related oxidative stress and cognitive decline | | Descriptor: | 8-methyl-2-(4-methyl-3-piperazin-1-ylsulfonyl-phenyl)imidazo[1,2-a]pyridine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Gould, N.L, Scherer, G.R, Carvalh, S, Shurrush, K, Edry, E, Elkobi, A, David, O, Dym, O, Albeck, S, Peleg, Y, Germain, N, Babaev, I, Sharir, H, Lefker, B, Subramanyam, C, Barr, H, Rosenblum, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Specific quinone reductase 2 inhibitors reduce metabolic burden and reverse Alzheimer's disease phenotype in mice.
J.Clin.Invest., 133, 2023
|
|
6ERE
 
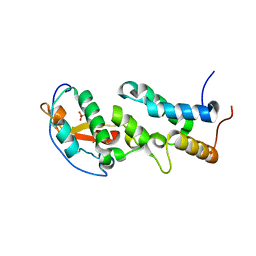 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes3/Imdes3 | | Descriptor: | Immunity, PHOSPHATE ION, colicin | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
6ER6
 
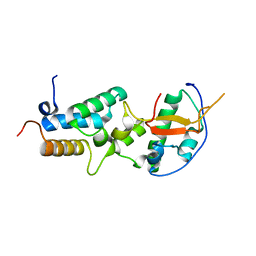 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes7/Imdes7 | | Descriptor: | Endonuclease colEdes7, immunity Imdes7 | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
5HQ3
 
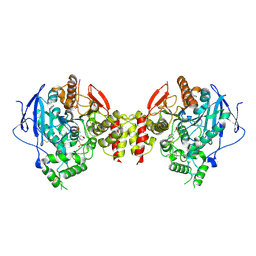 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
4FL0
 
 | | Crystal structure of ALD1 from Arabidopsis thaliana | | Descriptor: | Aminotransferase ALD1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sobolev, V, Edelman, M, Dym, O, Unger, T, Albeck, S, Kirma, M, Galili, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2012-06-14 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ALD1, a plant-specific homologue of the universal diaminopimelate aminotransferase enzyme of lysine biosynthesis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1XXM
 
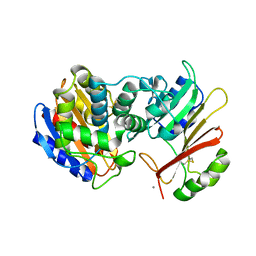 | | The modular architecture of protein-protein binding site | | Descriptor: | Beta-lactamase TEM, Beta-lactamase inhibitory protein, CALCIUM ION | | Authors: | Reichmann, D, Rahat, O, Albeck, S, Meged, R, Dym, O, Schreiber, G, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-11-07 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The modular architecture of protein-protein binding interfaces
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4BLF
 
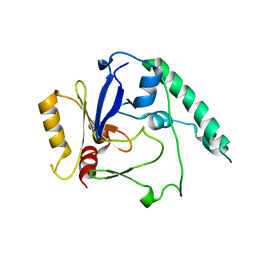 | | Variable internal flexibility characterizes the helical capsid formed by Agrobacterium VirE2 protein on single-stranded DNA. | | Descriptor: | SINGLE-STRAND DNA-BINDING PROTEIN | | Authors: | Bharat, T.A.M, Zbaida, D, Eisenstein, M, Frankenstein, Z, Mehlman, T, Weiner, L, Sorzano, C.O.S, Barak, Y, Albeck, S, Briggs, J.A.G, Wolf, S.G, Elbaum, M. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Variable Internal Flexibility Characterizes the Helical Capsid Formed by Agrobacterium Vire2 Protein on Single-Stranded DNA.
Structure, 21, 2013
|
|
3BTP
 
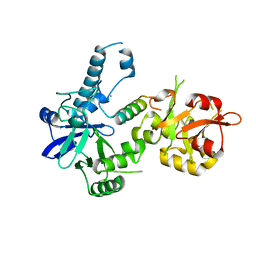 | | Crystal structure of Agrobacterium tumefaciens VirE2 in complex with its chaperone VirE1: a novel fold and implications for DNA binding | | Descriptor: | AMMONIUM ION, DI(HYDROXYETHYL)ETHER, Protein virE1, ... | | Authors: | Dym, O, Albeck, S, Unger, T, Elbaum, M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-12-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2YAT
 
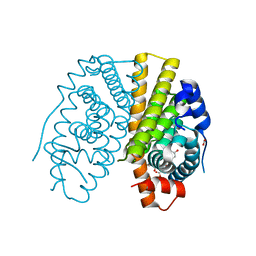 | | Crystal structure of estradiol derived metal chelate and estrogen receptor-ligand binding domain complex | | Descriptor: | ESTRADIOL-PYRIDINIUM TETRAACETIC ACID, ESTROGEN RECEPTOR, EUROPIUM ION, ... | | Authors: | Li, M.J, Greenblatt, H.M, Dym, O, Albeck, S, Degani, H, Sussman, J.L. | | Deposit date: | 2011-02-24 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure of Estradiol Metal Chelate and Estrogen Receptor Complex: The Basis for Designing a New Class of Selective Estrogen Receptor Modulators.
J.Med.Chem., 54, 2011
|
|
2RKX
 
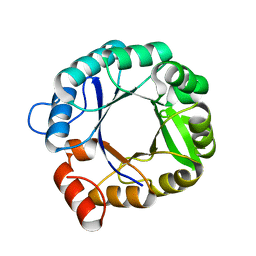 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Descriptor: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | Authors: | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-10-18 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
8P7I
 
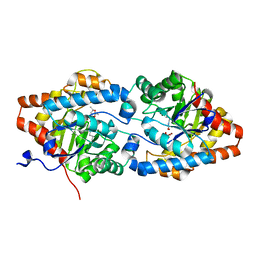 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | (2~{S},6~{R})-2,6-dimethyl-1,3-dioxane-4,4-diol, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7N
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | FORMIC ACID, Parathion hydrolase, ZINC ION | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7V
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggawal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7S
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7U
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7T
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-31 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7Q
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7R
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, FORMIC ACID, GLYCEROL, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7M
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | FORMIC ACID, GLYCEROL, METHYLPHOSPHONIC ACID ESTER GROUP, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7H
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | 1,2-ETHANEDIOL, 2-methylidene-1,3-dioxane-4,4-diol, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8P7F
 
 | | The impact of molecular variants, crystallization conditions and space group on structure-ligand complexes: A case study on Bacterial Phosphotriesterase Variants and complexes | | Descriptor: | Parathion hydrolase, ZINC ION | | Authors: | Dym, O, Aggarwal, N, Ashani, Y, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Sussman, J.L. | | Deposit date: | 2023-05-30 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of molecular variants, crystallization conditions and the space group on ligand-protein complexes: a case study on bacterial phosphotriesterase.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
