1CDN
 
 | |
1CB1
 
 | |
2XG3
 
 | | Human galectin-3 in complex with a benzamido-N-acetyllactoseamine inhibitor | | Descriptor: | BENZAMIDE, CHLORIDE ION, Galectin-3, ... | | Authors: | Diehl, C, Engstrom, O, Delaine, T, Hakansson, M, Genheden, S, Modig, K, Leffler, H, Ryde, U, Nilsson, U, Akke, M. | | Deposit date: | 2010-05-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3.
J. Am. Chem. Soc., 132, 2010
|
|
5EHB
 
 | | A de novo designed hexameric coiled-coil peptide with iodotyrosine | | Descriptor: | pHiosYI | | Authors: | Lizatovic, R, Aurelius, O, Stenstrom, O, Drakenberg, T, Akke, M, Logan, D.T, Andre, I. | | Deposit date: | 2015-10-28 | | Release date: | 2016-06-15 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | A De Novo Designed Coiled-Coil Peptide with a Reversible pH-Induced Oligomerization Switch.
Structure, 24, 2016
|
|
3ZSK
 
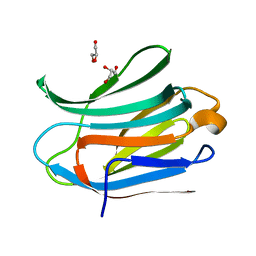 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | Descriptor: | GALECTIN-3, GLYCEROL | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
1CNP
 
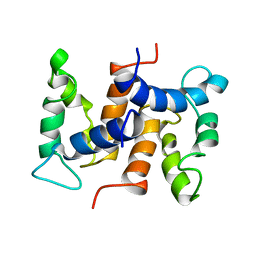 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
3ZSM
 
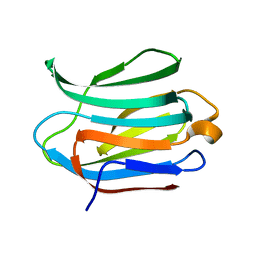 | | Crystal structure of Apo Human Galectin-3 CRD at 1.25 angstrom resolution, at room temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
2BCA
 
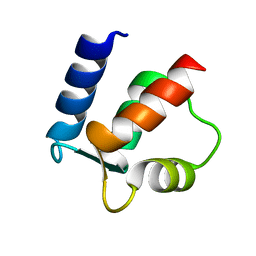 | |
2BCB
 
 | |
3ZSJ
 
 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSL
 
 | | Crystal structure of Apo Human Galectin-3 CRD at 1.08 angstrom resolution, at cryogenic temperature | | Descriptor: | GALECTIN-3 | | Authors: | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | Deposit date: | 2011-06-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
6EXY
 
 | | Neutron crystal structure of perdeuterated galectin-3C in complex with glycerol | | Descriptor: | GLYCEROL, Galectin-3 | | Authors: | Manzoni, F, Schrader, T.E, Ostermann, A, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-10 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
6EYM
 
 | | Neutron crystal structure of perdeuterated galectin-3C in complex with lactose | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Manzoni, F, Coates, L, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
1A03
 
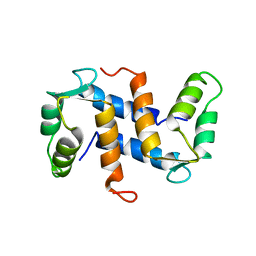 | | THE THREE-DIMENSIONAL STRUCTURE OF CA2+-BOUND CALCYCLIN: IMPLICATIONS FOR CA2+-SIGNAL TRANSDUCTION BY S100 PROTEINS, NMR, 20 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, CA2+) | | Authors: | Sastry, M, Ketchem, R.R, Crescenzi, O, Weber, C, Lubienski, M.J, Hidaka, H, Chazin, W.J. | | Deposit date: | 1997-12-08 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of Ca(2+)-bound calcyclin: implications for Ca(2+)-signal transduction by S100 proteins.
Structure, 6, 1998
|
|
1D1O
 
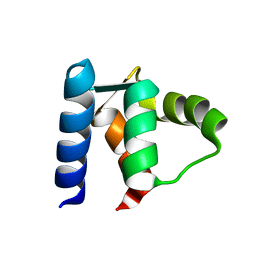 | |
2N1A
 
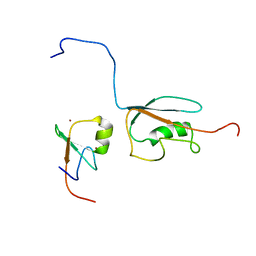 | | Docked structure between SUMO1 and ZZ-domain from CBP | | Descriptor: | CREB-binding protein, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Diehl, C. | | Deposit date: | 2015-03-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of a Complex between Small Ubiquitin-like Modifier 1 (SUMO1) and the ZZ Domain of CREB-binding Protein (CBP/p300) Reveals a New Interaction Surface on SUMO.
J.Biol.Chem., 291, 2016
|
|
6F2Q
 
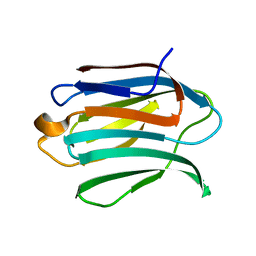 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
2CNP
 
 | |
5OAX
 
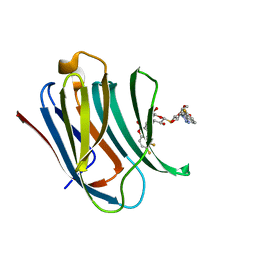 | | Galectin-3c in complex with thiogalactoside derivate | | Descriptor: | 5,6-bis(fluoranyl)-3-[[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]chromen-2-one, Galectin-3 | | Authors: | Nilsson, U.J, Peterson, K, Hakansson, M, Logan, D.T. | | Deposit date: | 2017-06-25 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity.
J. Med. Chem., 61, 2018
|
|
5NMS
 
 | | Hsp21 dodecamer, structural model based on cryo-EM and homology modelling | | Descriptor: | 25.3 kDa heat shock protein, chloroplastic | | Authors: | Rutsdottir, G, Harmark, J, Koeck, P.J.B, Hebert, H, Soderberg, C.A.G, Emanuelsson, C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity.
J. Biol. Chem., 292, 2017
|
|
5ODY
 
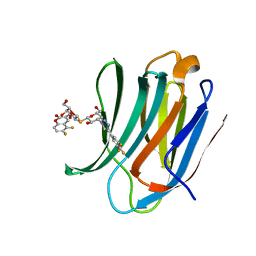 | | Galectin-3C in complex with dithiogalactoside derivative | | Descriptor: | 5,6-bis(fluoranyl)-3-[[(2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]chromen-2-one, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2017-07-07 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Systematic Tuning of Fluoro-galectin-3 Interactions Provides Thiodigalactoside Derivatives with Single-Digit nM Affinity and High Selectivity.
J. Med. Chem., 61, 2018
|
|
1JWD
 
 | |
6T86
 
 | | Urocanate reductase in complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T87
 
 | | Urocanate reductase in complex with urocanate | | Descriptor: | (2E)-3-(1H-IMIDAZOL-4-YL)ACRYLIC ACID, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Venskutonyte, R, Lindkvist-Petersson, K. | | Deposit date: | 2019-10-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural characterization of the microbial enzyme urocanate reductase mediating imidazole propionate production.
Nat Commun, 12, 2021
|
|
6T88
 
 | |
