2BF9
 
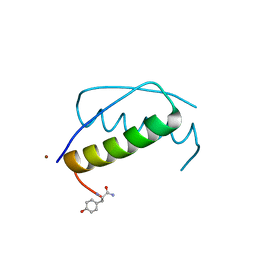 | | Anisotropic refinement of avian (turkey) pancreatic polypeptide at 0. 99 Angstroms resolution. | | Descriptor: | PANCREATIC HORMONE, ZINC ION | | Authors: | Tickle, I, Glover, I, Pitts, J, Wood, S, Blundell, T.L. | | Deposit date: | 2004-12-06 | | Release date: | 2004-12-08 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Conformational Flexibility in a Small Globular Hormone. X-Ray Analysis of Avian Pancreatic Polypeptide at 0.98 Angstroms Resolution
Biopolymers, 22, 1983
|
|
2BJI
 
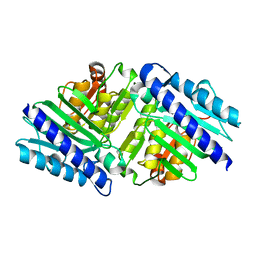 | | High Resolution Structure of myo-Inositol Monophosphatase, The Target of Lithium Therapy | | Descriptor: | INOSITOL-1(OR 4)-MONOPHOSPHATASE, MAGNESIUM ION | | Authors: | Gill, R, Mohammed, F, Badyal, R, Coates, L, Erskine, P, Thompson, D, Cooper, J, Gore, M, Wood, S. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | High-resolution structure of myo-inositol monophosphatase, the putative target of lithium therapy.
Acta Crystallogr. D Biol. Crystallogr., 61, 2005
|
|
7AVY
 
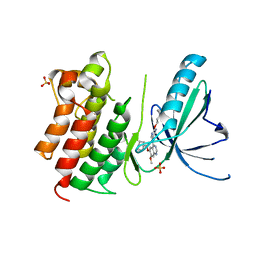 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVX
 
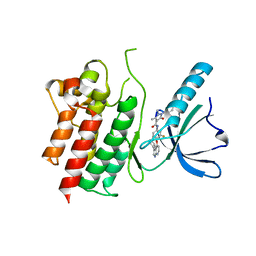 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW0
 
 | | MerTK kinase domain in complex with purine inhibitor | | Descriptor: | 2-(cyclopentyloxy)-9-(2,6-difluorobenzyl)-N-methyl-9H-purin-6-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Clarke, M, Disch, J, Goldberg, K, Guilinger, J, Hennessy, E.J, Jetson, R, Ginkunja, D, Hardaker, E, Keefe, A, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S, Zhang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW4
 
 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW1
 
 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVZ
 
 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW3
 
 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
3PSN
 
 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3PSO
 
 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3RSX
 
 | |
3RTM
 
 | |
3RU1
 
 | |
3RVI
 
 | |
1PPT
 
 | | X-RAY ANALYSIS (1.4-ANGSTROMS RESOLUTION) OF AVIAN PANCREATIC POLYPEPTIDE. SMALL GLOBULAR PROTEIN HORMONE | | Descriptor: | AVIAN PANCREATIC POLYPEPTIDE, ZINC ION | | Authors: | Blundell, T.L, Pitts, J.E, Tickle, I.J, Wood, S.P. | | Deposit date: | 1981-01-16 | | Release date: | 1981-02-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray analysis (1. 4-A resolution) of avian pancreatic polypeptide: Small globular protein hormone.
Proc.Natl.Acad.Sci.Usa, 78, 1981
|
|
6WNY
 
 | | Crystal structure of BACE1 in complex with (Z)-fluoro-olefin containing compound 15 | | Descriptor: | 6-[(Z)-2-{3-[(1S,5S,6S)-3-amino-5-methyl-1-(morpholine-4-carbonyl)-2-thia-4-azabicyclo[4.1.0]hept-3-en-5-yl]-4-fluorophenyl}-1-fluoroethenyl]pyridine-3-carbonitrile, Beta-secretase 1, IODIDE ION | | Authors: | Whittington, D.A. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The development of a structurally distinct series of BACE1 inhibitors via the (Z)-fluoro-olefin amide bioisosteric replacement.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5L9A
 
 | | L-threonine dehydrogenase from trypanosoma brucei. | | Descriptor: | ACETATE ION, L-threonine 3-dehydrogenase | | Authors: | Erskine, P.T, Cooper, J.B, Adjogatse, E, Kelly, J, Wood, S.P. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5LC1
 
 | | L-threonine dehydrogenase from Trypanosoma brucei with NAD and the inhibitor pyruvate bound. | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, L-threonine 3-dehydrogenase, ... | | Authors: | Erskine, P.T, Adjogatse, E, Wood, S.P, Cooper, J.B. | | Deposit date: | 2016-06-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of L-threonine-3-dehydrogenase from the parasitic protozoan Trypanosoma brucei revealed by X-ray crystallography and geometric simulations.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4WTU
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-aza-4-fluoro-xanthene inhibitor 22 | | Descriptor: | (5S)-3-(5,6-dihydro-2H-pyran-3-yl)-1-fluoro-7-(2-fluoropyridin-3-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Orally Available BACE1 Inhibitor That Affords Robust CNS A beta Reduction without Cardiovascular Liabilities.
Acs Med.Chem.Lett., 6, 2015
|
|
6B8O
 
 | | WT Ig-like V Domain with Phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sudom, A, Wang, Z. | | Deposit date: | 2017-10-09 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the loss-of-function effects of the Alzheimer's disease-associated R47H variant of the immune receptor TREM2.
J. Biol. Chem., 293, 2018
|
|
4XKX
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-azaxanthene inhibitor 28 | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-(2-fluoropyridin-4-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious beta-secretase inhibitors for the potential treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4RCE
 
 | | Crystal structure of BACE1 in complex with aminooxazoline xanthene inhibitor 2 | | Descriptor: | (4S)-2'-(2,2-dimethylpropoxy)-7'-(pyrimidin-5-yl)spiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lead Optimization and Modulation of hERG Activity in a Series of Aminooxazoline Xanthene beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4RCD
 
 | | Crystal structure of BACE1 in complex with a 2-aminooxazoline 4-azaxanthene inhibitor | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-[(3-methyloxetan-3-yl)ethynyl]spiro[chromeno[2,3-b]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2014-09-15 | | Release date: | 2015-01-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of beta-Site Amyloid Precursor Protein Cleaving Enzyme (BACE1): Identification of (S)-7-(2-Fluoropyridin-3-yl)-3-((3-methyloxetan-3-yl)ethynyl)-5'H-spiro[chromeno[2,3-b]pyridine-5,4'-oxazol]-2'-amine (AMG-8718).
J.Med.Chem., 57, 2014
|
|
