2PER
 
 | |
5EMC
 
 | | Transcription factor GRDBD and smGRE complex | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*(5CM)P*AP*TP*CP*AP*TP*GP*TP*TP*(5CM)P*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*(5CM)P*AP*TP*GP*AP*TP*GP*TP*TP*(5CM)P*TP*G)-3'), Glucocorticoid receptor, ... | | Authors: | Su, X.D, Lian, T, Jin, J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effects of cytosine methylation on general transcription factors
To Be Published
|
|
3V97
 
 | |
3V8V
 
 | |
1KTV
 
 | |
3JCU
 
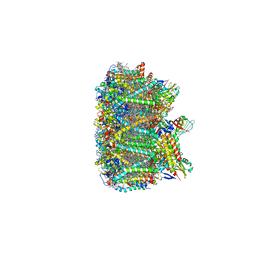 | | Cryo-EM structure of spinach PSII-LHCII supercomplex at 3.2 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Wei, X.P, Zhang, X.Z, Su, X.D, Cao, P, Liu, X.Y, Li, M, Chang, W.R, Liu, Z.F. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of spinach photosystem II-LHCII supercomplex at 3.2 A resolution
Nature, 534, 2016
|
|
3CZC
 
 | |
3H6X
 
 | |
5GRM
 
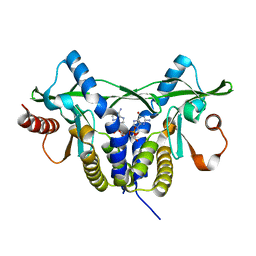 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
4E2A
 
 | |
1R3U
 
 | | Crystal Structure of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis | | Descriptor: | ACETATE ION, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Chen, Q, Liang, Y.H, Gu, X.C, Luo, M, Su, X.D. | | Deposit date: | 2003-10-03 | | Release date: | 2004-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Hypoxanthine-Guanine Phosphoribosyltransferase from Thermoanaerobacter tengcongensis
To be published
|
|
1RN7
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-11-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1PFR
 
 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (III) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-12-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1RKB
 
 | | The structure of adrenal gland protein AD-004 | | Descriptor: | LITHIUM ION, Protein AD-004, SULFATE ION | | Authors: | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | Deposit date: | 2003-11-21 | | Release date: | 2005-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ROA
 
 | | Structure of human cystatin D | | Descriptor: | Cystatin D | | Authors: | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
2RI1
 
 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
2RI0
 
 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucosamine-6-phosphate deaminase, SODIUM ION | | Authors: | Li, D, Liu, C, Li, L.F, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
2AYD
 
 | | Crystal Structure of the C-terminal WRKY domainof AtWRKY1, an SA-induced and partially NPR1-dependent transcription factor | | Descriptor: | SUCCINIC ACID, WRKY transcription factor 1, ZINC ION | | Authors: | Duan, M.R, Nan, J, Li, Y, Su, X.D. | | Deposit date: | 2005-09-07 | | Release date: | 2006-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein.
Nucleic Acids Res., 35, 2007
|
|
2B78
 
 | |
2BB0
 
 | |
7BUJ
 
 | | mcGAS bound with pppGpG | | Descriptor: | Cyclic GMP-AMP synthase, GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, B, Su, X.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mn2+Directly Activates cGAS and Structural Analysis Suggests Mn2+Induces a Noncanonical Catalytic Synthesis of 2'3'-cGAMP.
Cell Rep, 32, 2020
|
|
3BR8
 
 | |
3BGK
 
 | |
2D4G
 
 | |
