1HKG
 
 | |
2YHX
 
 | |
3HVT
 
 | | STRUCTURAL BASIS OF ASYMMETRY IN THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE HETERODIMER | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Steitz, T.A, Smerdon, S.J, Jaeger, J, Wang, J, Kohlstaedt, L.A, Chirino, A.J, Friedman, J.M, Rice, P.A. | | Deposit date: | 1994-07-25 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.Usa, 91, 1994
|
|
2ZIM
 
 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Steitz, T.A, Kavran, J.M. | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3HIF
 
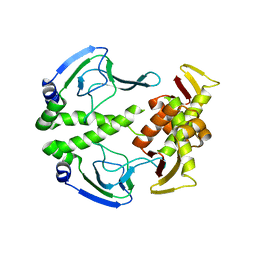 | | The crystal structure of apo wild type CAP at 3.6 A resolution. | | Descriptor: | Catabolite gene activator | | Authors: | Steitz, T.A, Sharma, H, Wang, J, Kong, J, Yu, S. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2QA4
 
 | |
2ODR
 
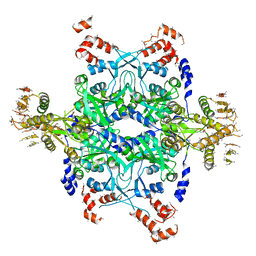 | | Methanococcus Maripaludis Phosphoseryl-tRNA synthetase | | Descriptor: | phosphoseryl-tRNA synthetase | | Authors: | Steitz, T.A, Kamtekar, S. | | Deposit date: | 2006-12-26 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.228 Å) | | Cite: | Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7JNH
 
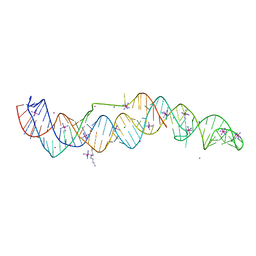 | | Crystal structure of a double-ENE RNA stability element in complex with a 28-mer poly(A) RNA | | Descriptor: | 28-mer poly(A) RNA, COBALT HEXAMMINE(III), Core double ENE RNA (Xtal construct) from Oryza sativa transposon,Core double ENE RNA (Xtal construct) from Oryza sativa transposon, ... | | Authors: | Torabi, S.F, Vaidya, A.T, Tycowski, K.T, DeGregorio, S.J, Wang, J, Shu, M.D, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2020-08-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Science, 371, 2021
|
|
4W2H
 
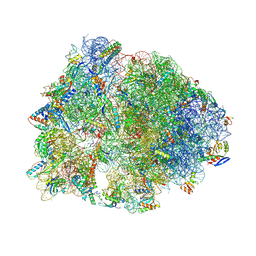 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
5TBZ
 
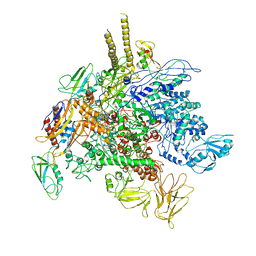 | | E. Coli RNA Polymerase complexed with NusG | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Steitz, T.A. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Structural insights into NusG regulating transcription elongation.
Nucleic Acids Res., 45, 2017
|
|
4Y4O
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome with rRNA modifications and bound to protein Y (YfiA) at 2.3A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Polikanov, Y.S, Melnikov, S.V, Soll, D, Steitz, T.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2KZZ
 
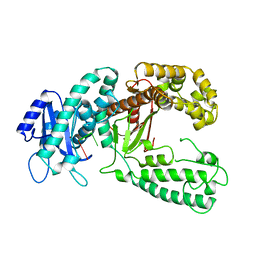 | | KLENOW FRAGMENT WITH NORMAL SUBSTRATE AND ZINC ONLY | | Descriptor: | DNA (5'-D(*GP*CP*TP*T*AP*CP*G)-3'), PROTEIN (DNA POLYMERASE I), ZINC ION | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-07 | | Release date: | 1999-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
1QU3
 
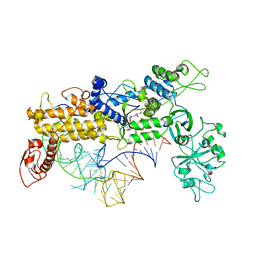 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MUPIROCIN, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
4PLX
 
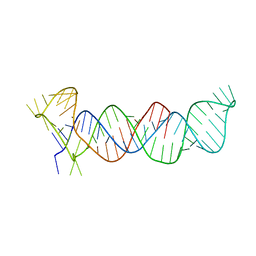 | | Crystal structure of the triple-helical stability element at the 3' end of MALAT1 | | Descriptor: | Core ENE hairpin and A-rich tract from MALAT1 | | Authors: | Brown, J.A, Bulkley, D, Wang, J, Valenstein, M.L, Yario, T.A, Steitz, T.A, Steitz, J.A. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1GSG
 
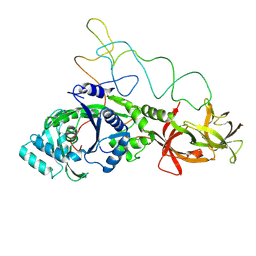 | | Structure of E.coli glutaminyl-tRNA synthetase complexed with trnagln and ATP at 2.8 Angstroms resolution | | Descriptor: | GLUTAMINYL-TRNA SYNTHETASE, TRNAGLN | | Authors: | Rould, M.A, Perona, J.J, Soell, D, Steitz, T.A. | | Deposit date: | 1990-04-03 | | Release date: | 1992-02-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution.
Science, 246, 1989
|
|
4M6F
 
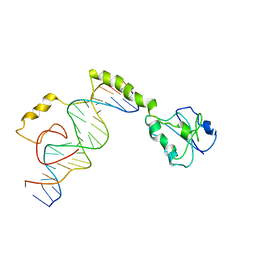 | |
4V8G
 
 | | Crystal structure of RMF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V5H
 
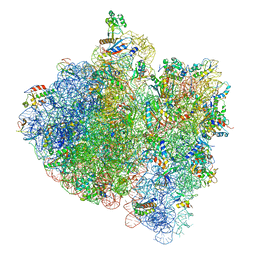 | | E.Coli 70s Ribosome Stalled During Translation Of Tnac Leader Peptide. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Seidelt, B, Innis, C.A, Wilson, D.N, Gartmann, M, Armache, J, Villa, E, Trabuco, L.G, Becker, T, Mielke, T, Schulten, K, Steitz, T.A, Beckmann, R. | | Deposit date: | 2009-10-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insight into nascent polypeptide chain-mediated translational stalling.
Science, 326, 2009
|
|
6VPR
 
 | |
6VPQ
 
 | |
1GTS
 
 | |
6B6H
 
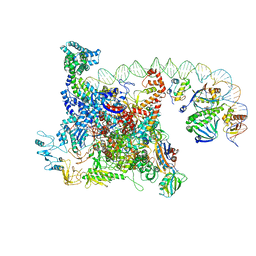 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
430D
 
 | | STRUCTURE OF SARCIN/RICIN LOOP FROM RAT 28S RRNA | | Descriptor: | MAGNESIUM ION, SARCIN/RICIN LOOP FROM RAT 28S R-RNA | | Authors: | Correll, C.C, Munishkin, A, Chan, Y.L, Ren, Z, Wool, I.G, Steitz, T.A. | | Deposit date: | 1998-10-04 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ribosomal RNA domain essential for binding elongation factors.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4W2F
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with amicoumacin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4W2G
 
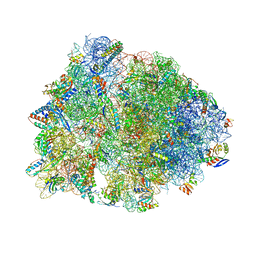 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (soaked), mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
