7UTF
 
 | | Structure-Function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol | | Descriptor: | CITRATE ANION, Putative oxidoreductase, aryl-alcohol dehydrogenase like protein, ... | | Authors: | Abraham, N, Schroeter, K.L, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function characterization of an aldo-keto reductase involved in detoxification of the mycotoxin, deoxynivalenol.
Sci Rep, 12, 2022
|
|
6OK1
 
 | | Ltp2-ChsH2(DUF35) aldolase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ChsH2(DUF35), ... | | Authors: | Kimber, M.S, Mallette, E, Aggett, R, Seah, S.Y.K. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The steroid side-chain-cleaving aldolase Ltp2-ChsH2DUF35is a thiolase superfamily member with a radically repurposed active site.
J.Biol.Chem., 294, 2019
|
|
6WY9
 
 | | Tcur3481-Tcur3483 steroid ACAD G363A variant | | Descriptor: | Acyl-CoA dehydrogenase domain protein Tcur3481, Acyl-CoA dehydrogenase domain protein Tcur3483, CHLORIDE ION, ... | | Authors: | Kimber, M.S, Stirling, A.J, Seah, S.Y.K. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity.
Biochemistry, 59, 2020
|
|
6WY8
 
 | | Tcur3481-Tcur3483 steroid ACAD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA dehydrogenase domain protein Tcur3481, Acyl-CoA dehydrogenase domain protein Tcur3483, ... | | Authors: | Kimber, M.S, Stirling, A.J, Seah, S.Y.K. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Glycine in Bacterial Steroid-Degrading Acyl-CoA Dehydrogenases Allows Flavin-Ring Repositioning and Modulates Substrate Side Chain Specificity.
Biochemistry, 59, 2020
|
|
4B5T
 
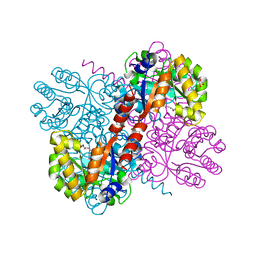 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5S
 
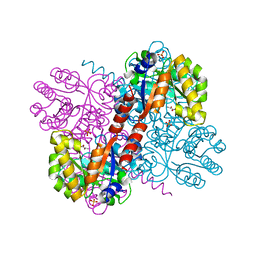 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, COBALT (II) ION, D-Glyceraldehyde, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5V
 
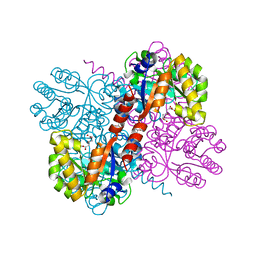 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with 4-hydroxyl-2-ketoheptane-1,7-dioate | | Descriptor: | (4R)-4-oxidanyl-2-oxidanylidene-heptanedioic acid, 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5U
 
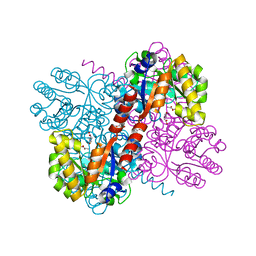 | | Crystal structures of divalent metal dependent pyruvate aldolase, HpaI, in complex with pyruvate and succinic semialdehyde | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, 4-oxobutanoic acid, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.913 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5W
 
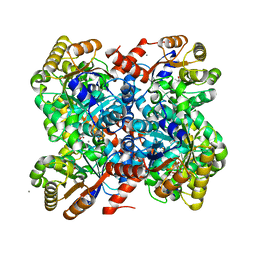 | | Crystal structures of divalent metal dependent pyruvate aldolase R70A mutant, HpaI, in complex with pyruvate | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
4B5X
 
 | | Crystal structures of divalent metal dependent pyruvate aldolase (HpaI), mutant D42A | | Descriptor: | 4-HYDROXY-2-OXO-HEPTANE-1,7-DIOATE ALDOLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Coincon, M, Wang, W, Seah, S.Y.K, Sygusch, J. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Reaction Intermediates in Pyruvate Class II Aldolase: Substrate Cleavage, Enolate Stabilization and Substrate Specificity
J.Biol.Chem., 287, 2012
|
|
5CGZ
 
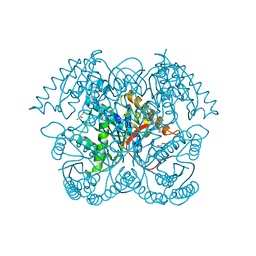 | | Crystal structure of GalB, the 4-carboxy-2-hydroxymuconate hydratase, from Pseuodomonas putida KT2440 | | Descriptor: | 4-oxalmesaconate hydratase, GLYCEROL, ZINC ION | | Authors: | Mazurkewich, S, Brott, A.S, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2015-07-09 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural and Kinetic Characterization of the 4-Carboxy-2-hydroxymuconate Hydratase from the Gallate and Protocatechuate 4,5-Cleavage Pathways of Pseudomonas putida KT2440.
J.Biol.Chem., 291, 2016
|
|
3NOJ
 
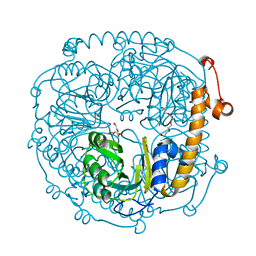 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | Descriptor: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | Deposit date: | 2010-06-25 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
4JN6
 
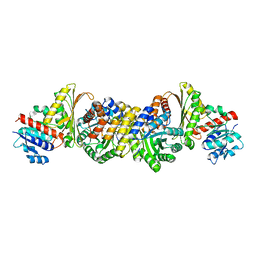 | | Crystal Structure of the Aldolase-Dehydrogenase Complex from Mycobacterium tuberculosis HRv37 | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, MANGANESE (II) ION, ... | | Authors: | Carere, J, McKenna, S.E, Kimber, M.S, Seah, S.Y.K. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of an Aldolase-Dehydrogenase Complex from the Cholesterol Degradation Pathway of Mycobacterium tuberculosis.
Biochemistry, 52, 2013
|
|
