3SAO
 
 | | The Siderocalin Ex-FABP functions through dual ligand specificities | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2,3-DIHYDROXY-BENZOIC ACID, Extracellular fatty acid-binding protein, ... | | Authors: | Correnti, C, Strong, R.K, Clifton, M.C. | | Deposit date: | 2011-06-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Galline Ex-FABP Is an Antibacterial Siderocalin and a Lysophosphatidic Acid Sensor Functioning through Dual Ligand Specificities.
Structure, 19, 2011
|
|
3U9P
 
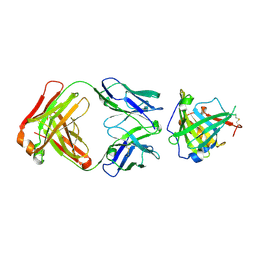 | | Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment | | Descriptor: | Monoclonal Fab Fragment Heavy Chain, Monoclonal Fab Fragment Light Chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Correnti, C, Strong, R.K. | | Deposit date: | 2011-10-19 | | Release date: | 2013-05-01 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines.
Plos One, 7, 2012
|
|
3S26
 
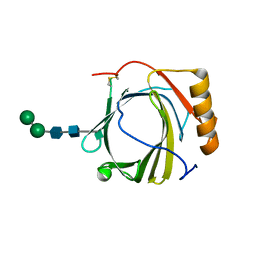 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
4K19
 
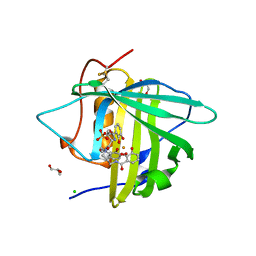 | | The structure of Human Siderocalin bound to the bacterial siderophore fluvibactin | | Descriptor: | (4S,5R)-N,N-bis{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, CHLORIDE ION, FE (III) ION, ... | | Authors: | Correnti, C, Clifton, M.C, Strong, R.K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Siderocalin Outwits the Coordination Chemistry of Vibriobactin, a Siderophore of Vibrio cholerae.
Acs Chem.Biol., 8, 2013
|
|
2KDK
 
 | |
6YAX
 
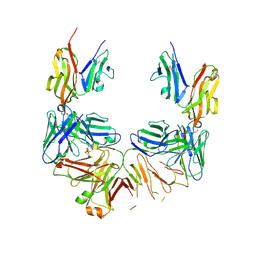 | |
7ZVX
 
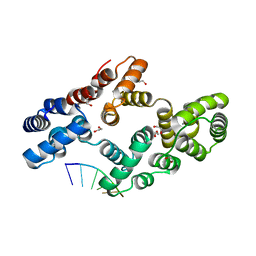 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7ZVN
 
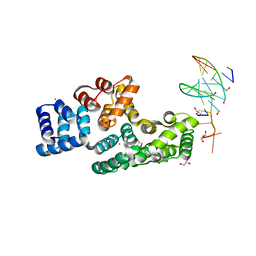 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 A resolution | | Descriptor: | 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, CALCIUM ION, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
5FTO
 
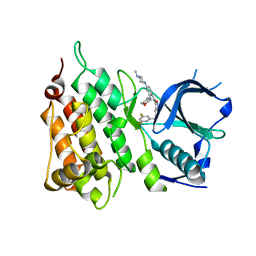 | | Crystal structure of the ALK kinase domain in complex with Entrectinib | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, Entrectinib | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
5FTQ
 
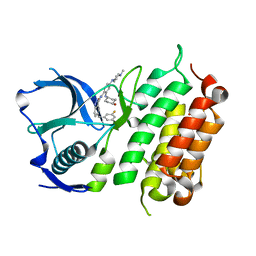 | | Crystal structure of the ALK kinase domain in complex with Cmpd 17 | | Descriptor: | ALK TYROSINE KINASE RECEPTOR, GLYCEROL, N-[5-(3,5-DIFLUOROBENZYL)-1H-INDAZOL-3-YL]-2-[(4-HYDROXYCYCLOHEXYL)AMINO]-4-(4-METHYLPIPERAZIN-1-YL) BENZAMIDE | | Authors: | Bossi, R, Canevari, G, Fasolini, M, Menichincheri, M, Ardini, E, Magnaghi, P, Avanzi, N, Banfi, P, Buffa, L, Ceriani, L, Colombo, M, Corti, L, Donati, D, Felder, E, Fiorelli, C, Fiorentini, F, Galvani, A, Isacchi, A, Lombardi Borgia, A, Marchionni, C, Nesi, M, Orrenius, C, Panzeri, A, Perrone, E, Pesenti, E, Rusconi, L, Saccardo, M.B, Vanotti, E, Orsini, P. | | Deposit date: | 2016-01-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Entrectinib: A New 3-Aminoindazole as a Potent Anaplastic Lymphoma Kinase (Alk), C-Ros Oncogene 1 Kinase (Ros1), and Pan-Tropomyosin Receptor Kinases (Pan-Trks) Inhibitor.
J.Med.Chem., 59, 2016
|
|
4L8I
 
 | | Crystal structure of RSV epitope scaffold FFL_005 | | Descriptor: | RSV epitope scaffold FFL_005 | | Authors: | Jardine, J, Correnti, C, Holmes, M.A, Strong, R.K, Schief, W.R. | | Deposit date: | 2013-06-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
2BKI
 
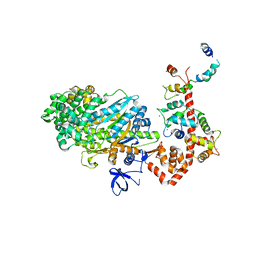 | | Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure | | Descriptor: | CALCIUM ION, CALMODULIN, SULFATE ION, ... | | Authors: | Menetrey, J, Bahloul, A, Yengo, C, Wells, A, Morris, C, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the Myosin Vi Motor Reveals the Mechanism of Directionality Reversal
Nature, 435, 2005
|
|
2BKH
 
 | | Myosin VI nucleotide-free (MDInsert2) crystal structure | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Menetrey, J, Bahloul, A, Yengo, C, Wells, A, Morris, C, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Myosin Vi Motor Reveals the Mechanism of Directionality Reversal
Nature, 435, 2005
|
|
6YAA
 
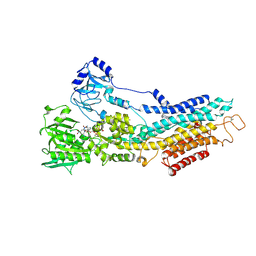 | | Structure of the (SR) Ca2+-ATPase bound to the inhibitor compound CAD204520 and TNP-ATP | | Descriptor: | 4-[2-[(2~{R})-2-[3-propyl-6-(trifluoromethyloxy)-1~{H}-indol-2-yl]piperidin-1-yl]ethyl]morpholine, POTASSIUM ION, SPIRO(2,4,6-TRINITROBENZENE[1,2A]-2O',3O'-METHYLENE-ADENINE-TRIPHOSPHATE, ... | | Authors: | Heit, S, Marchesini, M, Gherli, A, Montanaro, A, Patrizi, L, Sorrentino, C, Pagliaro, L, Rompietti, C, Kitara, S, Olesen, C.E, Moller, J.V, Savi, M, Bocchi, L, Vilella, R, Rizzi, F, Baglione, M, Rastelli, G, Loiacona, C, La Starza, R, Mecucci, C, Stegmair, K, Aversa, F, Stilli, D, Lund Winther, A.M, Sportoletti, P, Dalby-Brown, W, Roti, G, Bublitz, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blockade of Oncogenic NOTCH1 with the SERCA Inhibitor CAD204520 in T Cell Acute Lymphoblastic Leukemia.
Cell Chem Biol, 27, 2020
|
|
6DA1
 
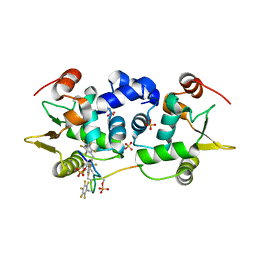 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.000127 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
6DAT
 
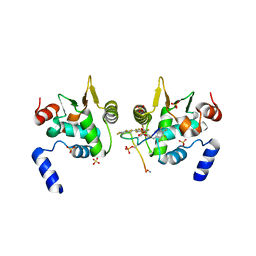 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35002637 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
8A38
 
 | |
8AMR
 
 | | Coiled-coil domain of human TRIM3 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Tripartite motif-containing protein 3 | | Authors: | Perez-Borrajero, C, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
4BZN
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo(1,2-a)Pyrazinone inhibitor | | Descriptor: | N-(2,2-dimethylpropyl)-2-[1-oxo-7-(thiophen-3-yl)-1,2,3,4-tetrahydropyrrolo[1,2-a]pyrazin-4-yl]acetamide, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
4BZO
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone inhibitor | | Descriptor: | N-[(1S)-2-AMINO-1-PHENYLETHYL]-2-[(4S)-7-(2-FLUORO-4-PYRIDINYL)-1-OXO-1,2,3,4-TETRAHYDROPYRROLO[1,2-A]PYRAZIN-4-YL]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
6ATY
 
 | |
6ATM
 
 | |
3U0D
 
 | |
5LLQ
 
 | |
